Process
mitochondrial genome maintenance (12 genes)
single strand break repair (9 genes)
ribosomal large subunit assembly (28 genes)
ribosomal small subunit assembly (15 genes)
very long-chain fatty acid metabolic process (17 genes)
autophagosome assembly (60 genes)
urea cycle (11 genes)
citrulline metabolic process (4 genes)
ribosomal large subunit export from nucleus (10 genes)
ribosomal small subunit export from nucleus (6 genes)
protein import into nucleus, translocation (23 genes)
mitotic sister chromatid segregation (29 genes)
dna replication checkpoint (5 genes)
dna damage checkpoint (29 genes)
regulation of cyclin-dependent protein serine/threonine kinase activity (35 genes)
g1/s transition of mitotic cell cycle (59 genes)
regulation of transcription involved in g1/s transition of mitotic cell cycle (7 genes)
g2/m transition of mitotic cell cycle (38 genes)
sulfate assimilation (4 genes)
negative regulation of transcription by rna polymerase ii (829 genes)
establishment of mitotic spindle orientation (25 genes)
actin cortical patch assembly (6 genes)
phosphorelay signal transduction system (5 genes)
mapk cascade (82 genes)
chromatin silencing at rdna (7 genes)
nuclear-transcribed mrna catabolic process, nonsense-mediated decay (32 genes)
activation of mapkkk activity (8 genes)
activation of mapkk activity (30 genes)
activation of mapk activity (83 genes)
inactivation of mapk activity (17 genes)
protein polyubiquitination (113 genes)
meiotic spindle organization (8 genes)
microtubule cytoskeleton organization (117 genes)
leptotene (6 genes)
spliceosomal tri-snrnp complex assembly (9 genes)
spliceosomal complex assembly (17 genes)
allantoin metabolic process (4 genes)
mitochondrial fission (17 genes)
mitotic cell cycle (124 genes)
mitotic cytokinesis (64 genes)
nuclear-transcribed mrna catabolic process, deadenylation-dependent decay (10 genes)
nuclear-transcribed mrna poly(a) tail shortening (11 genes)
deadenylation-dependent decapping of nuclear-transcribed mrna (10 genes)
response to reactive oxygen species (18 genes)
response to superoxide (5 genes)
re-entry into mitotic cell cycle (4 genes)
protein deneddylation (9 genes)
rna splicing, via transesterification reactions (8 genes)
alternative mrna splicing, via spliceosome (18 genes)
regulation of alternative mrna splicing, via spliceosome (64 genes)
spliceosomal snrnp assembly (23 genes)
mrna 5'-splice site recognition (8 genes)
mrna splicing, via spliceosome (109 genes)
protein peptidyl-prolyl isomerization (22 genes)
autophagy of mitochondrion (33 genes)
mitophagy (7 genes)
endonucleolytic cleavage in its1 to separate ssu-rrna from 5.8s rrna and lsu-rrna from tricistronic rrna transcript (ssu-rrna, 5.8s rrna, lsu-rrna) (7 genes)
snorna guided rrna pseudouridine synthesis (4 genes)
maturation of 5.8s rrna (7 genes)
maturation of ssu-rrna from tricistronic rrna transcript (ssu-rrna, 5.8s rrna, lsu-rrna) (24 genes)
maturation of lsu-rrna from tricistronic rrna transcript (ssu-rrna, 5.8s rrna, lsu-rrna) (12 genes)
maturation of 5.8s rrna from tricistronic rrna transcript (ssu-rrna, 5.8s rrna, lsu-rrna) (4 genes)
exonucleolytic trimming to generate mature 3'-end of 5.8s rrna from tricistronic rrna transcript (ssu-rrna, 5.8s rrna, lsu-rrna) (7 genes)
maturation of lsu-rrna (7 genes)
endonucleolytic cleavage in 5'-ets of tricistronic rrna transcript (ssu-rrna, 5.8s rrna, lsu-rrna) (4 genes)
box c/d snornp assembly (11 genes)
embryonic axis specification (9 genes)
meiotic dna repair synthesis (4 genes)
resolution of meiotic recombination intermediates (18 genes)
telomere maintenance via recombination (11 genes)
telomere maintenance (43 genes)
double-strand break repair via homologous recombination (74 genes)
double-strand break repair via break-induced replication (5 genes)
dna double-strand break processing (6 genes)
dna recombinase assembly (4 genes)
dna synthesis involved in dna repair (9 genes)
dna strand renaturation (5 genes)
dna catabolic process, endonucleolytic (11 genes)
dna catabolic process, exonucleolytic (5 genes)
sister chromatid segregation (4 genes)
cell morphogenesis (82 genes)
cell morphogenesis involved in differentiation (10 genes)
cytokinesis (5 genes)
actomyosin contractile ring assembly (5 genes)
cell separation after cytokinesis (7 genes)
nuclear-transcribed mrna catabolic process (18 genes)
mitochondrial mrna catabolic process (4 genes)
replicative cell aging (5 genes)
skeletal system development (103 genes)
cartilage condensation (19 genes)
ossification (99 genes)
neurotransmitter uptake (9 genes)
regulation of neurotransmitter levels (9 genes)
action potential (20 genes)
rna methylation (11 genes)
selenocysteine incorporation (6 genes)
prostaglandin biosynthetic process (15 genes)
pseudouridine synthesis (16 genes)
retinoid metabolic process (12 genes)
angiogenesis (240 genes)
ovarian follicle development (50 genes)
ovulation from ovarian follicle (9 genes)
antral ovarian follicle growth (4 genes)
luteinization (9 genes)
oocyte maturation (22 genes)
regulation of cell growth (60 genes)
fatty acid alpha-oxidation (5 genes)
blood vessel development (72 genes)
branching involved in blood vessel morphogenesis (40 genes)
vasculogenesis (71 genes)
ganglioside biosynthetic process (9 genes)
microtubule bundle formation (32 genes)
detection of chemical stimulus involved in sensory perception of bitter taste (48 genes)
osteoblast differentiation (67 genes)
eye development (37 genes)
urogenital system development (16 genes)
metanephros development (38 genes)
ureteric bud development (46 genes)
branching involved in ureteric bud morphogenesis (46 genes)
temperature homeostasis (22 genes)
fever generation (4 genes)
conditioned taste aversion (5 genes)
behavioral fear response (42 genes)
response to hypoxia (124 genes)
ameboidal-type cell migration (9 genes)
acrosome assembly (14 genes)
long-chain fatty acid metabolic process (21 genes)
cellular glucose homeostasis (21 genes)
trna 5'-leader removal (11 genes)
gastric acid secretion (9 genes)
in utero embryonic development (287 genes)
gastrulation with mouth forming second (27 genes)
endoderm formation (12 genes)
mesoderm formation (34 genes)
cell fate specification (22 genes)
cell fate determination (18 genes)
endodermal cell fate commitment (7 genes)
endodermal cell fate specification (5 genes)
formation of translation preinitiation complex (10 genes)
establishment of planar polarity (21 genes)
morphogenesis of a polarized epithelium (6 genes)
neural crest cell migration (50 genes)
somitogenesis (63 genes)
somite specification (4 genes)
organ induction (15 genes)
morphogenesis of a branching structure (11 genes)
neuron migration (133 genes)
membrane raft assembly (7 genes)
establishment of t cell polarity (5 genes)
immunological synapse formation (10 genes)
myeloid dendritic cell activation (4 genes)
microglial cell activation (20 genes)
cell activation (10 genes)
leukocyte homeostasis (9 genes)
plasma membrane repair (8 genes)
natural killer cell differentiation (17 genes)
neutrophil homeostasis (7 genes)
b cell homeostasis (28 genes)
b cell apoptotic process (13 genes)
positive regulation of type iia hypersensitivity (4 genes)
positive regulation of type iii hypersensitivity (4 genes)
positive regulation of type i hypersensitivity (4 genes)
cytokine production (30 genes)
regulation of cytokine production (13 genes)
negative regulation of cytokine production (14 genes)
positive regulation of cytokine production (32 genes)
histamine secretion (6 genes)
kidney development (111 genes)
mesonephros development (10 genes)
blastocyst development (31 genes)
blastocyst formation (29 genes)
inner cell mass cell differentiation (4 genes)
inner cell mass cell fate commitment (4 genes)
inner cell mass cellular morphogenesis (4 genes)
trophectodermal cell differentiation (21 genes)
blastocyst growth (10 genes)
inner cell mass cell proliferation (16 genes)
blastocyst hatching (40 genes)
release of cytochrome c from mitochondria (21 genes)
epithelial to mesenchymal transition (32 genes)
neural plate morphogenesis (4 genes)
neural tube formation (18 genes)
neural fold formation (5 genes)
neural tube closure (100 genes)
phagolysosome assembly (4 genes)
nk t cell differentiation (4 genes)
complement activation, lectin pathway (9 genes)
response to yeast (4 genes)
mullerian duct regression (4 genes)
receptor recycling (11 genes)
endothelial cell development (5 genes)
endothelial cell morphogenesis (16 genes)
selenium compound metabolic process (4 genes)
liver development (71 genes)
placenta development (48 genes)
embryonic placenta development (29 genes)
maternal placenta development (11 genes)
tissue homeostasis (25 genes)
retina homeostasis (13 genes)
t cell mediated cytotoxicity (23 genes)
positive regulation of t cell mediated cytotoxicity (45 genes)
regulation of receptor recycling (8 genes)
negative regulation of receptor recycling (4 genes)
positive regulation of receptor recycling (17 genes)
b-1 b cell homeostasis (5 genes)
regulation of protein phosphorylation (71 genes)
negative regulation of protein phosphorylation (100 genes)
positive regulation of protein phosphorylation (208 genes)
endothelial cell proliferation (13 genes)
regulation of endothelial cell proliferation (5 genes)
negative regulation of endothelial cell proliferation (33 genes)
positive regulation of endothelial cell proliferation (70 genes)
hair follicle development (53 genes)
vasculature development (27 genes)
lymph vessel development (13 genes)
lymphangiogenesis (15 genes)
heart looping (57 genes)
regulation of cell-matrix adhesion (13 genes)
negative regulation of cell-matrix adhesion (19 genes)
positive regulation of cell-matrix adhesion (28 genes)
blood vessel maturation (7 genes)
positive regulation of neurotransmitter secretion (8 genes)
intramembranous ossification (5 genes)
endochondral ossification (28 genes)
negative regulation of cytokine-mediated signaling pathway (8 genes)
positive regulation of cytokine-mediated signaling pathway (8 genes)
synaptic transmission, dopaminergic (20 genes)
startle response (21 genes)
suckling behavior (17 genes)
adenosine receptor signaling pathway (8 genes)
blood vessel remodeling (42 genes)
response to amphetamine (19 genes)
regulation of systemic arterial blood pressure by circulatory renin-angiotensin (5 genes)
regulation of systemic arterial blood pressure by vasopressin (5 genes)
norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure (4 genes)
angiotensin-mediated vasoconstriction involved in regulation of systemic arterial blood pressure (4 genes)
angiotensin maturation (4 genes)
morphogenesis of an epithelium (27 genes)
morphogenesis of an epithelial sheet (12 genes)
renin-angiotensin regulation of aldosterone production (4 genes)
response to dietary excess (15 genes)
reduction of food intake in response to dietary excess (6 genes)
diet induced thermogenesis (13 genes)
regulation of the force of heart contraction (24 genes)
regulation of heart rate (32 genes)
regulation of sodium ion transport (23 genes)
desensitization of g-protein coupled receptor protein signaling pathway (9 genes)
g-protein coupled receptor internalization (12 genes)
brain renin-angiotensin system (6 genes)
sprouting angiogenesis (31 genes)
cell migration involved in sprouting angiogenesis (22 genes)
blood vessel endothelial cell proliferation involved in sprouting angiogenesis (7 genes)
osteoblast fate commitment (7 genes)
positive regulation of neuroblast proliferation (27 genes)
positive regulation of mesenchymal cell proliferation (36 genes)
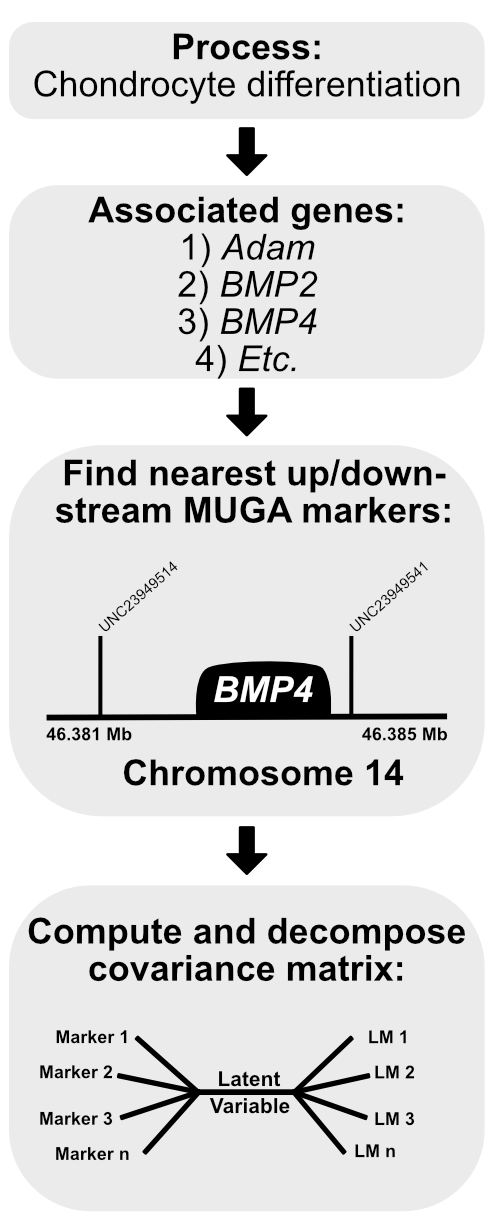
chondrocyte differentiation (38 genes)
chondrocyte development (19 genes)
epithelial cell development (13 genes)
epithelial cell maturation (8 genes)
osteoblast development (20 genes)
regulation of oxidative phosphorylation (11 genes)
protein depalmitoylation (9 genes)
regulation of respiratory gaseous exchange by neurological system process (13 genes)
lens development in camera-type eye (34 genes)
lens morphogenesis in camera-type eye (16 genes)
regulation of receptor internalization (8 genes)
negative regulation of receptor internalization (8 genes)
positive regulation of receptor internalization (28 genes)
auditory receptor cell morphogenesis (6 genes)
trna wobble uridine modification (14 genes)
store-operated calcium entry (8 genes)
aggressive behavior (4 genes)
retinoic acid biosynthetic process (5 genes)
trna wobble position uridine thiolation (4 genes)
regulation of thyroid hormone mediated signaling pathway (5 genes)
desmosome assembly (4 genes)
protein localization to paranode region of axon (5 genes)
cytoplasmic translation (35 genes)
translation reinitiation (4 genes)
defense response to nematode (4 genes)
activation of innate immune response (22 genes)
toll-like receptor signaling pathway (18 genes)
innate immune response in mucosa (41 genes)
positive regulation of defense response to virus by host (30 genes)
leukocyte chemotaxis involved in inflammatory response (4 genes)
response to molecule of bacterial origin (12 genes)
hematopoietic progenitor cell differentiation (95 genes)
connective tissue replacement involved in inflammatory response wound healing (5 genes)
adaptive immune response (142 genes)
lymphocyte homeostasis (12 genes)
myeloid cell homeostasis (11 genes)
macrophage activation involved in immune response (12 genes)
neutrophil activation involved in immune response (5 genes)
t cell activation involved in immune response (29 genes)
t cell differentiation involved in immune response (4 genes)
t-helper 1 cell lineage commitment (4 genes)
t cell proliferation involved in immune response (4 genes)
marginal zone b cell differentiation (10 genes)
plasma cell differentiation (4 genes)
myeloid progenitor cell differentiation (8 genes)
lymphoid progenitor cell differentiation (7 genes)
b cell proliferation involved in immune response (5 genes)
natural killer cell activation involved in immune response (26 genes)
b cell lineage commitment (5 genes)
pro-b cell differentiation (5 genes)
pre-b cell differentiation (10 genes)
t cell lineage commitment (6 genes)
cd4-positive, cd25-positive, alpha-beta regulatory t cell differentiation (4 genes)
cytokine secretion involved in immune response (7 genes)
immune system process (444 genes)
immunoglobulin production (13 genes)
mucosal immune response (5 genes)
dendritic cell chemotaxis (5 genes)
myeloid dendritic cell chemotaxis (4 genes)
complement receptor mediated signaling pathway (13 genes)
inflammatory response to antigenic stimulus (16 genes)
acute inflammatory response to antigenic stimulus (7 genes)
neutrophil mediated immunity (9 genes)
humoral immune response mediated by circulating immunoglobulin (4 genes)
t cell mediated immunity (7 genes)
t cell antigen processing and presentation (5 genes)
germinal center formation (7 genes)
antigen processing and presentation of peptide antigen via mhc class i (8 genes)
antigen processing and presentation of endogenous peptide antigen via mhc class ib (30 genes)
antigen processing and presentation of peptide or polysaccharide antigen via mhc class ii (5 genes)
immune system development (4 genes)
leukocyte differentiation (4 genes)
leukocyte migration involved in inflammatory response (13 genes)
acute inflammatory response (15 genes)
monocyte chemotaxis (33 genes)
mast cell chemotaxis (5 genes)
histamine secretion by mast cell (5 genes)
myeloid leukocyte differentiation (6 genes)
positive regulation of dendritic cell antigen processing and presentation (6 genes)
regulation of germinal center formation (4 genes)
positive regulation of germinal center formation (5 genes)
positive regulation of immunoglobulin production (11 genes)
positive regulation of t cell tolerance induction (4 genes)
positive regulation of t cell anergy (4 genes)
regulation of acute inflammatory response (4 genes)
negative regulation of acute inflammatory response (8 genes)
positive regulation of acute inflammatory response (8 genes)
negative regulation of chronic inflammatory response (4 genes)
regulation of immune system process (5 genes)
negative regulation of leukocyte migration (6 genes)
positive regulation of leukocyte migration (22 genes)
positive regulation of leukocyte chemotaxis (9 genes)
positive regulation of cellular extravasation (7 genes)
negative regulation of t cell cytokine production (6 genes)
positive regulation of t cell cytokine production (8 genes)
regulation of dendritic cell cytokine production (4 genes)
negative regulation of cytokine secretion involved in immune response (5 genes)
positive regulation of cytokine secretion involved in immune response (9 genes)
myd88-dependent toll-like receptor signaling pathway (15 genes)
myd88-independent toll-like receptor signaling pathway (4 genes)
positive regulation of peptide secretion (4 genes)
regulation of adaptive immune response (7 genes)
positive regulation of adaptive immune response (9 genes)
positive regulation of t-helper 1 type immune response (12 genes)
negative regulation of type 2 immune response (7 genes)
positive regulation of type 2 immune response (8 genes)
positive regulation of immune response to tumor cell (6 genes)
positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target (7 genes)
negative regulation of inflammatory response to antigenic stimulus (9 genes)
negative regulation of acute inflammatory response to antigenic stimulus (4 genes)
negative regulation of b cell apoptotic process (18 genes)
positive regulation of b cell apoptotic process (5 genes)
negative regulation of mature b cell apoptotic process (5 genes)
positive regulation of humoral immune response mediated by circulating immunoglobulin (6 genes)
response to ischemia (29 genes)
desmosome organization (5 genes)
developmental process involved in reproduction (4 genes)
heart morphogenesis (66 genes)
skeletal muscle contraction (20 genes)
renal system process (10 genes)
respiratory system process (14 genes)
regulation of vascular smooth muscle contraction (4 genes)
regulation of systemic arterial blood pressure (27 genes)
regulation of systemic arterial blood pressure by renin-angiotensin (5 genes)
positive regulation of systemic arterial blood pressure (11 genes)
negative regulation of systemic arterial blood pressure (17 genes)
renal water homeostasis (4 genes)
glomerular filtration (9 genes)
regulation of systemic arterial blood pressure by endothelin (5 genes)
positive regulation of glomerular filtration (4 genes)
negative regulation of glomerular filtration (5 genes)
secondary heart field specification (9 genes)
embryonic heart tube morphogenesis (10 genes)
outflow tract septum morphogenesis (29 genes)
membranous septum morphogenesis (10 genes)
muscular septum morphogenesis (5 genes)
outflow tract morphogenesis (55 genes)
endocardium development (5 genes)
cardiac conduction system development (6 genes)
heart valve development (11 genes)
aortic valve development (4 genes)
heart valve morphogenesis (8 genes)
aortic valve morphogenesis (17 genes)
atrioventricular valve morphogenesis (15 genes)
mitral valve morphogenesis (8 genes)
pulmonary valve morphogenesis (15 genes)
tricuspid valve morphogenesis (4 genes)
mitral valve formation (5 genes)
tricuspid valve formation (4 genes)
endocardial cushion development (14 genes)
epithelial to mesenchymal transition involved in endocardial cushion formation (12 genes)
endocardial cushion to mesenchymal transition involved in heart valve formation (4 genes)
endocardial cushion morphogenesis (18 genes)
cardiac ventricle morphogenesis (9 genes)
cardiac atrium morphogenesis (7 genes)
cardiac ventricle formation (5 genes)
cardiac left ventricle morphogenesis (14 genes)
cardiac right ventricle morphogenesis (12 genes)
cardiac right ventricle formation (4 genes)
ventricular trabecula myocardium morphogenesis (16 genes)
ventricular compact myocardium morphogenesis (8 genes)
ventricular cardiac muscle tissue development (6 genes)
cardiac ventricle development (7 genes)
cardiac neural crest cell migration involved in outflow tract morphogenesis (4 genes)
regulation of membrane depolarization (9 genes)
positive regulation of transcription from rna polymerase ii promoter involved in myocardial precursor cell differentiation (4 genes)
regulation of secondary heart field cardioblast proliferation (4 genes)
endocardial cushion formation (6 genes)
cell migration involved in endocardial cushion formation (4 genes)
endocardial cushion fusion (4 genes)
cardiac septum development (22 genes)
ventricular septum development (40 genes)
atrial septum development (6 genes)
atrial septum primum morphogenesis (5 genes)
cardiac muscle hypertrophy (8 genes)
type b pancreatic cell differentiation (10 genes)
pancreatic a cell differentiation (5 genes)
type b pancreatic cell development (16 genes)
amino acid transmembrane transport (27 genes)
keratinocyte development (12 genes)
mesenchymal to epithelial transition involved in metanephros morphogenesis (6 genes)
metanephros morphogenesis (4 genes)
cilium movement (32 genes)
pericardium morphogenesis (8 genes)
epithelial cilium movement (17 genes)
regulation of cilium beat frequency (5 genes)
noradrenergic neuron differentiation (4 genes)
dynamin family protein polymerization involved in mitochondrial fission (4 genes)
sphingosine-1-phosphate signaling pathway (5 genes)
epithelial cell morphogenesis (27 genes)
apical constriction (4 genes)
regulation of copii vesicle coating (6 genes)
axis elongation (4 genes)
retinal pigment epithelium development (6 genes)
neural retina development (7 genes)
chondrocyte differentiation involved in endochondral bone morphogenesis (5 genes)
endochondral bone growth (8 genes)
growth plate cartilage development (11 genes)
growth plate cartilage chondrocyte morphogenesis (16 genes)
detection of calcium ion (8 genes)
carbohydrate metabolic process (182 genes)
glycogen metabolic process (38 genes)
glycogen biosynthetic process (18 genes)
regulation of glycogen biosynthetic process (10 genes)
glycogen catabolic process (10 genes)
regulation of glycogen catabolic process (6 genes)
fructose metabolic process (7 genes)
fructose 6-phosphate metabolic process (9 genes)
fructose 2,6-bisphosphate metabolic process (6 genes)
fucose metabolic process (6 genes)
glucose metabolic process (62 genes)
glucose catabolic process (6 genes)
galactose metabolic process (7 genes)
mannose metabolic process (8 genes)
5-phosphoribose 1-diphosphate biosynthetic process (7 genes)
inositol metabolic process (8 genes)
glycosaminoglycan biosynthetic process (15 genes)
glycosaminoglycan catabolic process (4 genes)
proteoglycan metabolic process (6 genes)
chitin catabolic process (8 genes)
n-acetylglucosamine metabolic process (16 genes)
udp-n-acetylglucosamine metabolic process (6 genes)
udp-n-acetylglucosamine biosynthetic process (6 genes)
n-acetylneuraminate metabolic process (4 genes)
ethanol catabolic process (5 genes)
ethanol oxidation (4 genes)
glycerol metabolic process (13 genes)
glycerol-3-phosphate metabolic process (8 genes)
cellular aldehyde metabolic process (8 genes)
organic acid metabolic process (51 genes)
acetyl-coa metabolic process (9 genes)
acetyl-coa biosynthetic process (5 genes)
acetyl-coa biosynthetic process from pyruvate (5 genes)
lactate metabolic process (6 genes)
pyruvate metabolic process (10 genes)
generation of precursor metabolites and energy (12 genes)
gluconeogenesis (24 genes)
glycolytic process (31 genes)
pentose-phosphate shunt (8 genes)
tricarboxylic acid cycle (31 genes)
citrate metabolic process (8 genes)
isocitrate metabolic process (6 genes)
2-oxoglutarate metabolic process (16 genes)
succinyl-coa metabolic process (6 genes)
succinate metabolic process (10 genes)
oxaloacetate metabolic process (14 genes)
malate metabolic process (7 genes)
regulation of glycolytic process (9 genes)
regulation of gluconeogenesis (13 genes)
energy reserve metabolic process (11 genes)
nadh oxidation (5 genes)
oxidative phosphorylation (6 genes)
mitochondrial electron transport, nadh to ubiquinone (17 genes)
mitochondrial electron transport, succinate to ubiquinone (4 genes)
mitochondrial electron transport, ubiquinol to cytochrome c (14 genes)
mitochondrial electron transport, cytochrome c to oxygen (12 genes)
nucleobase-containing compound metabolic process (28 genes)
purine nucleobase metabolic process (7 genes)
purine nucleotide metabolic process (4 genes)
purine nucleotide biosynthetic process (21 genes)
nucleoside diphosphate phosphorylation (14 genes)
purine ribonucleoside salvage (4 genes)
amp biosynthetic process (5 genes)
camp biosynthetic process (11 genes)
cgmp biosynthetic process (11 genes)
gtp biosynthetic process (10 genes)
'de novo' imp biosynthetic process (4 genes)
'de novo' pyrimidine nucleobase biosynthetic process (6 genes)
pyrimidine nucleotide metabolic process (4 genes)
pyrimidine nucleotide biosynthetic process (7 genes)
dump biosynthetic process (4 genes)
utp biosynthetic process (8 genes)
dtmp biosynthetic process (4 genes)
ctp biosynthetic process (9 genes)
dna metabolic process (16 genes)
dna replication (122 genes)
dna-dependent dna replication (22 genes)
mitochondrial dna replication (11 genes)
dna topological change (8 genes)
dna ligation (5 genes)
dna unwinding involved in dna replication (9 genes)
dna replication, synthesis of rna primer (4 genes)
dna replication initiation (25 genes)
regulation of dna replication (17 genes)
dna repair (339 genes)
regulation of dna repair (6 genes)
transcription-coupled nucleotide-excision repair (10 genes)
base-excision repair (32 genes)
base-excision repair, gap-filling (5 genes)
nucleotide-excision repair (37 genes)
pyrimidine dimer repair (5 genes)
nucleotide-excision repair, dna gap filling (5 genes)
mismatch repair (20 genes)
postreplication repair (9 genes)
double-strand break repair (63 genes)
double-strand break repair via nonhomologous end joining (26 genes)
dna methylation (22 genes)
dna dealkylation involved in dna repair (6 genes)
dna catabolic process (7 genes)
apoptotic dna fragmentation (10 genes)
dna recombination (84 genes)
mitotic recombination (6 genes)
dna packaging (4 genes)
chromatin organization (328 genes)
chromatin assembly or disassembly (8 genes)
nucleosome assembly (86 genes)
dna replication-dependent nucleosome assembly (31 genes)
dna replication-independent nucleosome assembly (23 genes)
nucleosome disassembly (15 genes)
chromatin remodeling (87 genes)
chromatin silencing (17 genes)
maintenance of chromatin silencing (4 genes)
methylation-dependent chromatin silencing (10 genes)
regulation of gene expression by genetic imprinting (17 genes)
transcription, dna-templated (113 genes)
dna-templated transcription, initiation (19 genes)
dna-templated transcription, termination (10 genes)
regulation of transcription, dna-templated (1009 genes)
regulation of transcription by rna polymerase ii (475 genes)
transcription by rna polymerase i (7 genes)
transcription initiation from rna polymerase i promoter (4 genes)
termination of rna polymerase i transcription (4 genes)
rrna processing (134 genes)
transcription by rna polymerase ii (132 genes)
transcription initiation from rna polymerase ii promoter (33 genes)
transcription elongation from rna polymerase ii promoter (22 genes)
termination of rna polymerase ii transcription (7 genes)
7-methylguanosine mrna capping (7 genes)
mrna splice site selection (20 genes)
mrna polyadenylation (29 genes)
mrna cleavage (5 genes)
adenosine to inosine editing (7 genes)
transcription by rna polymerase iii (12 genes)
transcription initiation from rna polymerase iii promoter (4 genes)
trna splicing, via endonucleolytic cleavage and ligation (10 genes)
mitochondrial transcription (11 genes)
rna processing (61 genes)
mrna processing (340 genes)
mrna 3'-end processing by stem-loop binding and cleavage (5 genes)
trna modification (15 genes)
rna catabolic process (19 genes)
mrna catabolic process (31 genes)
rna export from nucleus (5 genes)
mrna export from nucleus (36 genes)
translation (259 genes)
translational initiation (54 genes)
translational elongation (20 genes)
translational termination (6 genes)
regulation of translation (131 genes)
trna aminoacylation for protein translation (34 genes)
regulation of translational initiation (20 genes)
regulation of translational fidelity (6 genes)
protein folding (89 genes)
cellular protein modification process (44 genes)
signal peptide processing (13 genes)
protein phosphorylation (579 genes)
negative regulation of protein kinase activity (66 genes)
protein dephosphorylation (150 genes)
protein adp-ribosylation (24 genes)
protein acetylation (13 genes)
n-terminal protein amino acid acetylation (9 genes)
internal protein amino acid acetylation (4 genes)
protein deacetylation (12 genes)
protein sulfation (4 genes)
protein methylation (29 genes)
protein demethylation (4 genes)
protein glycosylation (74 genes)
protein n-linked glycosylation (42 genes)
dolichol-linked oligosaccharide biosynthetic process (6 genes)
oligosaccharide-lipid intermediate biosynthetic process (5 genes)
n-glycan processing (12 genes)
protein o-linked glycosylation (30 genes)
protein lipidation (11 genes)
n-terminal protein myristoylation (4 genes)
gpi anchor biosynthetic process (31 genes)
proteolysis (574 genes)
membrane protein ectodomain proteolysis (20 genes)
ubiquitin-dependent protein catabolic process (258 genes)
protein monoubiquitination (35 genes)
protein quality control for misfolded or incompletely synthesized proteins (10 genes)
glycoprotein catabolic process (7 genes)
protein deglycosylation (8 genes)
peptide metabolic process (14 genes)
cellular amino acid metabolic process (22 genes)
arginine metabolic process (4 genes)
arginine biosynthetic process (6 genes)
arginine catabolic process (6 genes)
aspartate metabolic process (6 genes)
glutamate metabolic process (14 genes)
glutamate biosynthetic process (4 genes)
glutamine metabolic process (17 genes)
glycine metabolic process (9 genes)
histidine metabolic process (5 genes)
histidine catabolic process (4 genes)
isoleucine metabolic process (4 genes)
leucine catabolic process (4 genes)
methionine metabolic process (5 genes)
s-adenosylmethionine biosynthetic process (4 genes)
l-phenylalanine catabolic process (7 genes)
proline metabolic process (4 genes)
proline biosynthetic process (4 genes)
proline catabolic process (4 genes)
l-serine metabolic process (5 genes)
l-serine biosynthetic process (4 genes)
l-serine catabolic process (4 genes)
threonine catabolic process (6 genes)
tryptophan catabolic process (6 genes)
tyrosine catabolic process (5 genes)
valine metabolic process (5 genes)
melanin metabolic process (4 genes)
catecholamine metabolic process (8 genes)
thyroid hormone generation (8 genes)
polyamine metabolic process (7 genes)
polyamine biosynthetic process (9 genes)
creatine metabolic process (5 genes)
protein targeting (39 genes)
protein import into nucleus (72 genes)
nls-bearing protein import into nucleus (13 genes)
ribosomal protein import into nucleus (7 genes)
protein export from nucleus (35 genes)
protein targeting to membrane (39 genes)
srp-dependent cotranslational protein targeting to membrane (10 genes)
srp-dependent cotranslational protein targeting to membrane, translocation (10 genes)
posttranslational protein targeting to endoplasmic reticulum membrane (11 genes)
protein retention in er lumen (7 genes)
protein targeting to lysosome (16 genes)
protein targeting to vacuole (8 genes)
protein targeting to peroxisome (12 genes)
protein targeting to mitochondrion (19 genes)
protein processing involved in protein targeting to mitochondrion (6 genes)
lipid metabolic process (477 genes)
fatty acid metabolic process (167 genes)
fatty acid biosynthetic process (68 genes)
fatty acid beta-oxidation (42 genes)
unsaturated fatty acid biosynthetic process (6 genes)
acyl-coa metabolic process (31 genes)
triglyceride metabolic process (38 genes)
triglyceride mobilization (6 genes)
phospholipid metabolic process (45 genes)
phosphatidylethanolamine biosynthetic process (9 genes)
glycerophospholipid metabolic process (6 genes)
diacylglycerol biosynthetic process (4 genes)
phosphatidic acid biosynthetic process (11 genes)
phosphatidylcholine biosynthetic process (13 genes)
phosphatidylserine metabolic process (5 genes)
phosphatidylserine biosynthetic process (4 genes)
phosphatidylinositol biosynthetic process (13 genes)
glycerol ether metabolic process (4 genes)
sphingolipid metabolic process (39 genes)
sphingosine metabolic process (6 genes)
ceramide metabolic process (16 genes)
galactosylceramide biosynthetic process (4 genes)
sphingomyelin metabolic process (4 genes)
sphingomyelin catabolic process (5 genes)
sphingomyelin biosynthetic process (7 genes)
glycosphingolipid metabolic process (8 genes)
ganglioside catabolic process (7 genes)
leukotriene metabolic process (8 genes)
prostaglandin metabolic process (22 genes)
steroid biosynthetic process (60 genes)
cholesterol biosynthetic process (29 genes)
bile acid biosynthetic process (11 genes)
c21-steroid hormone biosynthetic process (4 genes)
androgen biosynthetic process (5 genes)
estrogen biosynthetic process (6 genes)
glucocorticoid biosynthetic process (6 genes)
cholesterol catabolic process (9 genes)
androgen catabolic process (4 genes)
cellular aromatic compound metabolic process (5 genes)
tetrahydrobiopterin biosynthetic process (7 genes)
one-carbon metabolic process (19 genes)
nadh metabolic process (14 genes)
nadp metabolic process (9 genes)
nadp biosynthetic process (5 genes)
nadp catabolic process (5 genes)
ubiquinone biosynthetic process (15 genes)
glutathione metabolic process (43 genes)
glutathione biosynthetic process (9 genes)
glutathione catabolic process (6 genes)
atp biosynthetic process (24 genes)
mo-molybdopterin cofactor biosynthetic process (6 genes)
porphyrin-containing compound metabolic process (5 genes)
porphyrin-containing compound biosynthetic process (13 genes)
heme biosynthetic process (18 genes)
sulfur compound metabolic process (18 genes)
phosphate-containing compound metabolic process (9 genes)
polyphosphate catabolic process (6 genes)
superoxide metabolic process (20 genes)
xenobiotic metabolic process (68 genes)
nitrogen compound metabolic process (21 genes)
nitric oxide biosynthetic process (10 genes)
transport (6 genes)
ion transport (603 genes)
cation transport (81 genes)
potassium ion transport (132 genes)
sodium ion transport (127 genes)
calcium ion transport (145 genes)
phosphate ion transport (10 genes)
anion transport (22 genes)
chloride transport (80 genes)
cobalt ion transport (6 genes)
copper ion transport (12 genes)
iron ion transport (13 genes)
manganese ion transport (14 genes)
zinc ii ion transport (28 genes)
water transport (16 genes)
neurotransmitter transport (44 genes)
mitochondrial transport (14 genes)
acyl carnitine transport (4 genes)
mitochondrial calcium ion transmembrane transport (13 genes)
drug transmembrane transport (13 genes)
oligopeptide transport (7 genes)
amino acid transport (47 genes)
glutamine transport (4 genes)
lipid transport (112 genes)
cellular ion homeostasis (4 genes)
cellular calcium ion homeostasis (98 genes)
cellular copper ion homeostasis (16 genes)
cellular iron ion homeostasis (45 genes)
intracellular sequestering of iron ion (11 genes)
cellular zinc ion homeostasis (20 genes)
cellular sodium ion homeostasis (19 genes)
cell volume homeostasis (24 genes)
regulation of ph (25 genes)
intracellular protein transport (320 genes)
exocytosis (110 genes)
er to golgi vesicle-mediated transport (110 genes)
retrograde vesicle-mediated transport, golgi to er (39 genes)
intra-golgi vesicle-mediated transport (25 genes)
post-golgi vesicle-mediated transport (10 genes)
golgi to plasma membrane transport (25 genes)
golgi to endosome transport (11 genes)
golgi to vacuole transport (7 genes)
endocytosis (178 genes)
receptor-mediated endocytosis (56 genes)
vesicle budding from membrane (8 genes)
vesicle targeting (6 genes)
vesicle docking involved in exocytosis (30 genes)
vesicle fusion (31 genes)
pinocytosis (4 genes)
phagocytosis (58 genes)
phagocytosis, recognition (12 genes)
phagocytosis, engulfment (28 genes)
nucleocytoplasmic transport (32 genes)
autophagy (160 genes)
apoptotic process (564 genes)
activation of cysteine-type endopeptidase activity involved in apoptotic process (80 genes)
activation-induced cell death of t cells (6 genes)
inflammatory cell apoptotic process (4 genes)
movement of cell or subcellular component (14 genes)
substrate-dependent cell migration (6 genes)
substrate-dependent cell migration, cell extension (8 genes)
negative regulation of cell adhesion involved in substrate-bound cell migration (4 genes)
chemotaxis (130 genes)
muscle contraction (48 genes)
regulation of muscle contraction (22 genes)
smooth muscle contraction (14 genes)
regulation of smooth muscle contraction (12 genes)
striated muscle contraction (13 genes)
regulation of striated muscle contraction (4 genes)
response to stress (4 genes)
defense response (113 genes)
acute-phase response (29 genes)
inflammatory response (324 genes)
immune response (240 genes)
complement activation (10 genes)
complement activation, alternative pathway (10 genes)
complement activation, classical pathway (29 genes)
humoral immune response (43 genes)
cellular defense response (14 genes)
response to osmotic stress (15 genes)
hyperosmotic response (5 genes)
cellular response to dna damage stimulus (466 genes)
dna damage induced protein phosphorylation (6 genes)
dna damage response, signal transduction by p53 class mediator resulting in cell cycle arrest (11 genes)
dna damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator (13 genes)
response to oxidative stress (123 genes)
er overload response (11 genes)
response to unfolded protein (63 genes)
cellular response to nitrogen starvation (10 genes)
organelle organization (14 genes)
nucleus organization (23 genes)
nuclear envelope organization (21 genes)
nuclear pore organization (7 genes)
nucleolus organization (6 genes)
telomere maintenance via telomerase (18 genes)
mitochondrion organization (81 genes)
mitochondrial membrane organization (7 genes)
inner mitochondrial membrane organization (7 genes)
plasma membrane organization (16 genes)
cytoskeleton organization (108 genes)
actin filament organization (105 genes)
cytoskeletal anchoring at plasma membrane (6 genes)
microtubule-based process (36 genes)
microtubule-based movement (80 genes)
microtubule depolymerization (16 genes)
microtubule nucleation (17 genes)
tubulin complex assembly (7 genes)
post-chaperonin tubulin folding pathway (4 genes)
negative regulation of microtubule depolymerization (33 genes)
cytoplasm organization (5 genes)
endoplasmic reticulum organization (28 genes)
golgi organization (97 genes)
peroxisome organization (24 genes)
endosome organization (40 genes)
vacuolar transport (15 genes)
vacuolar acidification (11 genes)
lysosome organization (40 genes)
lysosomal transport (11 genes)
lysosomal lumen acidification (8 genes)
cell-cell junction assembly (38 genes)
cell-substrate junction assembly (5 genes)
cell cycle (626 genes)
cell cycle arrest (86 genes)
spindle organization (19 genes)
mitotic spindle organization (35 genes)
chromosome segregation (87 genes)
sister chromatid cohesion (17 genes)
mitotic sister chromatid cohesion (13 genes)
mitotic chromosome condensation (14 genes)
mitotic metaphase plate congression (34 genes)
regulation of mitotic nuclear division (33 genes)
metaphase/anaphase transition of mitotic cell cycle (6 genes)
mitotic cell cycle checkpoint (30 genes)
mitotic spindle assembly checkpoint (25 genes)
mitotic g2 dna damage checkpoint (20 genes)
regulation of exit from mitosis (7 genes)
nuclear migration (16 genes)
centrosome cycle (39 genes)
centriole replication (19 genes)
mitotic centrosome separation (6 genes)
meiosis i (5 genes)
synapsis (32 genes)
synaptonemal complex assembly (22 genes)
reciprocal meiotic recombination (24 genes)
male meiotic nuclear division (32 genes)
male meiosis i (22 genes)
female meiotic nuclear division (12 genes)
female meiosis i (6 genes)
cell communication (37 genes)
cell adhesion (534 genes)
homophilic cell adhesion via plasma membrane adhesion molecules (109 genes)
heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules (46 genes)
neuron cell-cell adhesion (9 genes)
leukocyte cell-cell adhesion (24 genes)
cell-matrix adhesion (80 genes)
negative regulation of cell adhesion (49 genes)
establishment or maintenance of cell polarity (39 genes)
signal transduction (1103 genes)
cell surface receptor signaling pathway (184 genes)
enzyme linked receptor protein signaling pathway (6 genes)
receptor guanylyl cyclase signaling pathway (12 genes)
transmembrane receptor protein tyrosine kinase signaling pathway (82 genes)
activation of transmembrane receptor protein tyrosine kinase activity (5 genes)
epidermal growth factor receptor signaling pathway (41 genes)
negative regulation of epidermal growth factor-activated receptor activity (14 genes)
regulation of epidermal growth factor-activated receptor activity (4 genes)
transmembrane receptor protein serine/threonine kinase signaling pathway (22 genes)
transforming growth factor beta receptor signaling pathway (79 genes)
common-partner smad protein phosphorylation (6 genes)
smad protein complex assembly (7 genes)
g-protein coupled receptor signaling pathway (1674 genes)
g-protein coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger (44 genes)
adenylate cyclase-modulating g-protein coupled receptor signaling pathway (55 genes)
adenylate cyclase-activating g-protein coupled receptor signaling pathway (96 genes)
activation of adenylate cyclase activity (21 genes)
adenylate cyclase-activating dopamine receptor signaling pathway (11 genes)
adenylate cyclase-inhibiting g-protein coupled receptor signaling pathway (46 genes)
negative regulation of adenylate cyclase activity (14 genes)
adenylate cyclase-inhibiting dopamine receptor signaling pathway (11 genes)
adenylate cyclase-inhibiting g-protein coupled glutamate receptor signaling pathway (4 genes)
adenylate cyclase-inhibiting g-protein coupled acetylcholine receptor signaling pathway (7 genes)
adenylate cyclase-inhibiting serotonin receptor signaling pathway (4 genes)
g-protein coupled receptor signaling pathway coupled to cgmp nucleotide second messenger (5 genes)
phospholipase c-activating g-protein coupled receptor signaling pathway (53 genes)
activation of phospholipase c activity (17 genes)
positive regulation of cytosolic calcium ion concentration (166 genes)
protein kinase c-activating g-protein coupled receptor signaling pathway (24 genes)
serotonin receptor signaling pathway (6 genes)
dopamine receptor signaling pathway (15 genes)
g-protein coupled acetylcholine receptor signaling pathway (8 genes)
gamma-aminobutyric acid signaling pathway (27 genes)
glutamate receptor signaling pathway (6 genes)
g-protein coupled glutamate receptor signaling pathway (16 genes)
tachykinin receptor signaling pathway (6 genes)
neuropeptide signaling pathway (80 genes)
notch signaling pathway (121 genes)
notch receptor processing (10 genes)
positive regulation of transcription of notch receptor target (6 genes)
wnt signaling pathway, calcium modulating pathway (4 genes)
smoothened signaling pathway (72 genes)
integrin-mediated signaling pathway (73 genes)
i-kappab kinase/nf-kappab signaling (37 genes)
activation of nf-kappab-inducing kinase activity (16 genes)
i-kappab phosphorylation (12 genes)
cytoplasmic sequestering of nf-kappab (6 genes)
jnk cascade (32 genes)
activation of jnkk activity (7 genes)
activation of jun kinase activity (39 genes)
jun phosphorylation (4 genes)
jak-stat cascade (24 genes)
tyrosine phosphorylation of stat protein (10 genes)
nitric oxide mediated signal transduction (13 genes)
small gtpase mediated signal transduction (87 genes)
ras protein signal transduction (45 genes)
rho protein signal transduction (73 genes)
cell-cell signaling (62 genes)
chemical synaptic transmission (185 genes)
neurotransmitter secretion (29 genes)
neuron-neuron synaptic transmission (10 genes)
synaptic transmission, cholinergic (22 genes)
neuromuscular synaptic transmission (32 genes)
multicellular organism development (1053 genes)
gamete generation (20 genes)
germ cell development (41 genes)
spermatogenesis (437 genes)
spermatid development (125 genes)
sperm axoneme assembly (17 genes)
spermatid nucleus differentiation (8 genes)
spermatid nucleus elongation (5 genes)
female gamete generation (8 genes)
vitellogenesis (4 genes)
single fertilization (69 genes)
binding of sperm to zona pellucida (38 genes)
acrosome reaction (16 genes)
penetration of zona pellucida (9 genes)
fusion of sperm to egg plasma membrane involved in single fertilization (14 genes)
regulation of mitotic cell cycle (39 genes)
determination of left/right symmetry (80 genes)
gastrulation (37 genes)
segment specification (6 genes)
compartment pattern specification (4 genes)
pattern specification process (68 genes)
ectoderm development (9 genes)
nervous system development (391 genes)
neuroblast proliferation (19 genes)
negative regulation of neuroblast proliferation (8 genes)
axonogenesis (97 genes)
axon guidance (146 genes)
axon target recognition (4 genes)
axonal fasciculation (23 genes)
synapse assembly (51 genes)
central nervous system development (77 genes)
brain development (158 genes)
peripheral nervous system development (22 genes)
sensory organ development (6 genes)
salivary gland morphogenesis (9 genes)
hindgut morphogenesis (4 genes)
endoderm development (41 genes)
mesoderm development (26 genes)
mesodermal cell fate specification (4 genes)
heart development (246 genes)
adult heart development (16 genes)
muscle organ development (67 genes)
skeletal muscle tissue development (58 genes)
myoblast fusion (24 genes)
somatic muscle development (4 genes)
neuromuscular junction development (36 genes)
sex determination (11 genes)
sex differentiation (14 genes)
female pregnancy (55 genes)
embryo implantation (52 genes)
parturition (5 genes)
aging (50 genes)
cell aging (84 genes)
response to nutrient (22 genes)
respiratory gaseous exchange (24 genes)
digestion (15 genes)
excretion (16 genes)
body fluid secretion (4 genes)
lactation (32 genes)
blood coagulation (72 genes)
hemostasis (46 genes)
visual perception (136 genes)
phototransduction (24 genes)
phototransduction, visible light (4 genes)
sensory perception of sound (253 genes)
sensory perception of chemical stimulus (26 genes)
sensory perception of smell (1127 genes)
behavior (16 genes)
learning or memory (60 genes)
learning (67 genes)
memory (95 genes)
short-term memory (14 genes)
long-term memory (38 genes)
mating behavior (10 genes)
negative regulation of female receptivity (4 genes)
circadian rhythm (77 genes)
grooming behavior (15 genes)
locomotory behavior (107 genes)
adult walking behavior (45 genes)
feeding behavior (28 genes)
chemosensory behavior (6 genes)
blood circulation (10 genes)
regulation of heart contraction (25 genes)
trna processing (79 genes)
motor neuron axon guidance (27 genes)
mitochondrial fusion (13 genes)
toll signaling pathway (6 genes)
regulation of actin polymerization or depolymerization (11 genes)
establishment of blood-nerve barrier (4 genes)
mesodermal cell migration (4 genes)
axo-dendritic transport (10 genes)
anterograde axonal transport (29 genes)
retrograde axonal transport (10 genes)
protein localization (87 genes)
biological_process (4332 genes)
metabolic process (176 genes)
actin polymerization or depolymerization (12 genes)
negative regulation of dna replication (16 genes)
steroid metabolic process (93 genes)
cholesterol metabolic process (88 genes)
bile acid metabolic process (12 genes)
c21-steroid hormone metabolic process (10 genes)
androgen metabolic process (13 genes)
estrogen metabolic process (12 genes)
glucocorticoid metabolic process (6 genes)
regulation of blood pressure (52 genes)
cell death (37 genes)
opsonization (5 genes)
sulfate transport (17 genes)
regulation of g-protein coupled receptor protein signaling pathway (33 genes)
cell proliferation (224 genes)
positive regulation of cell proliferation (546 genes)
negative regulation of cell proliferation (394 genes)
insulin receptor signaling pathway (56 genes)
spermidine biosynthetic process (4 genes)
isoprenoid biosynthetic process (15 genes)
associative learning (37 genes)
endosome to lysosome transport (43 genes)
determination of adult lifespan (26 genes)
adult feeding behavior (10 genes)
adult locomotory behavior (70 genes)
glial cell migration (12 genes)
germ cell migration (9 genes)
olfactory learning (4 genes)
regulation of cell shape (145 genes)
regulation of cell size (23 genes)
axon ensheathment (6 genes)
rna splicing (254 genes)
gonad development (10 genes)
respiratory chain complex iv assembly (5 genes)
visual learning (67 genes)
fibroblast growth factor receptor signaling pathway (41 genes)
epidermis development (52 genes)
regulation of synaptic growth at neuromuscular junction (5 genes)
male gonad development (80 genes)
female gonad development (26 genes)
regulation of smoothened signaling pathway (25 genes)
regulation of notch signaling pathway (17 genes)
photoreceptor cell morphogenesis (8 genes)
anterior/posterior axis specification, embryo (6 genes)
attachment of spindle microtubules to kinetochore (9 genes)
lipid biosynthetic process (25 genes)
ether lipid biosynthetic process (5 genes)
extrinsic apoptotic signaling pathway via death domain receptors (27 genes)
granzyme-mediated apoptotic signaling pathway (11 genes)
intrinsic apoptotic signaling pathway in response to dna damage (56 genes)
intrinsic apoptotic signaling pathway in response to oxidative stress (17 genes)
activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c (6 genes)
apoptotic mitochondrial changes (27 genes)
carbohydrate transport (35 genes)
cellular amino acid biosynthetic process (24 genes)
phospholipid biosynthetic process (60 genes)
dosage compensation by inactivation of x chromosome (15 genes)
pentose-phosphate shunt, non-oxidative branch (5 genes)
biosynthetic process (38 genes)
aerobic respiration (31 genes)
fatty acid catabolic process (10 genes)
cellular amino acid catabolic process (4 genes)
aromatic amino acid family metabolic process (10 genes)
branched-chain amino acid catabolic process (9 genes)
methionine biosynthetic process (11 genes)
glycoprotein metabolic process (10 genes)
glycoprotein biosynthetic process (11 genes)
purine nucleobase biosynthetic process (4 genes)
xanthine catabolic process (5 genes)
nucleoside metabolic process (15 genes)
nucleotide metabolic process (27 genes)
nucleoside triphosphate biosynthetic process (9 genes)
nucleoside triphosphate catabolic process (6 genes)
ribonucleoside monophosphate biosynthetic process (5 genes)
nucleotide biosynthetic process (9 genes)
nucleotide catabolic process (4 genes)
purine ribonucleoside monophosphate biosynthetic process (6 genes)
cyclic nucleotide biosynthetic process (18 genes)
cobalamin metabolic process (6 genes)
glycolipid biosynthetic process (8 genes)
protein lipoylation (6 genes)
peptidoglycan catabolic process (6 genes)
deoxyribonucleotide catabolic process (4 genes)
response to temperature stimulus (7 genes)
cellular response to starvation (65 genes)
response to ph (13 genes)
mrna transcription (7 genes)
rrna transcription (13 genes)
protein secretion (38 genes)
amine metabolic process (9 genes)
oligosaccharide metabolic process (10 genes)
oligosaccharide biosynthetic process (12 genes)
oligosaccharide catabolic process (8 genes)
response to radiation (20 genes)
phospholipid catabolic process (17 genes)
toxin metabolic process (10 genes)
response to heat (46 genes)
response to cold (31 genes)
response to xenobiotic stimulus (8 genes)
response to uv (48 genes)
response to light stimulus (20 genes)
nad biosynthetic process (10 genes)
carnitine metabolic process (8 genes)
rna modification (13 genes)
fertilization (52 genes)
detection of virus (4 genes)
response to wounding (74 genes)
response to mechanical stimulus (27 genes)
response to virus (54 genes)
response to bacterium (248 genes)
response to fungus (6 genes)
response to toxic substance (54 genes)
entrainment of circadian clock (4 genes)
uv protection (10 genes)
response to salt stress (8 genes)
anatomical structure morphogenesis (41 genes)
response to hormone (39 genes)
response to carbohydrate (5 genes)
response to glucose (48 genes)
hormone-mediated signaling pathway (41 genes)
embryo development (55 genes)
post-embryonic development (116 genes)
embryo development ending in birth or egg hatching (19 genes)
coumarin metabolic process (5 genes)
abscission (4 genes)
embryonic pattern specification (20 genes)
animal organ morphogenesis (104 genes)
tissue development (18 genes)
auditory receptor cell fate commitment (4 genes)
epidermal cell differentiation (10 genes)
anterior/posterior axis specification (19 genes)
dorsal/ventral axis specification (10 genes)
anterior/posterior pattern specification (115 genes)
dorsal/ventral pattern formation (47 genes)
proximal/distal pattern formation (26 genes)
epidermal cell fate specification (4 genes)
regulation of signal transduction (36 genes)
positive regulation of signal transduction (14 genes)
negative regulation of signal transduction (111 genes)
cytidine deamination (4 genes)
cellular process (25 genes)
oocyte differentiation (4 genes)
glial cell differentiation (21 genes)
meiotic chromosome condensation (6 genes)
response to organic substance (70 genes)
response to inorganic substance (9 genes)
response to metal ion (7 genes)
response to iron ion (7 genes)
response to iron(ii) ion (4 genes)
response to manganese ion (5 genes)
response to zinc ion (20 genes)
proline catabolic process to glutamate (4 genes)
regulation of proton transport (5 genes)
response to x-ray (17 genes)
embryonic body morphogenesis (10 genes)
response to ionizing radiation (39 genes)
maintenance of dna methylation (8 genes)
response to uv-b (9 genes)
response to uv-c (9 genes)
response to organonitrogen compound (14 genes)
multicellular organism aging (16 genes)
response to selenium ion (6 genes)
detoxification of copper ion (6 genes)
response to lead ion (10 genes)
regulation of hydrogen peroxide metabolic process (4 genes)
membrane invagination (6 genes)
response to gamma radiation (24 genes)
negative regulation of anion channel activity (5 genes)
regulation of g2/m transition of mitotic cell cycle (13 genes)
histone monoubiquitination (12 genes)
response to acidic ph (9 genes)
histone h3-k36 methylation (4 genes)
regulation of cell fate commitment (5 genes)
negative regulation of cell fate commitment (4 genes)
centriole-centriole cohesion (12 genes)
exit from mitosis (11 genes)
negative regulation of heart rate (9 genes)
positive regulation of heart rate (23 genes)
mesenchymal cell proliferation (5 genes)
negative regulation of peptidase activity (118 genes)
gene expression (15 genes)
regulation of gene expression (333 genes)
regulation of receptor activity (18 genes)
regulation of gastrulation (11 genes)
proteasomal protein catabolic process (33 genes)
proteasomal ubiquitin-independent protein catabolic process (23 genes)
rna secondary structure unwinding (8 genes)
regulation of autophagy (58 genes)
negative regulation of autophagy (37 genes)
positive regulation of autophagy (47 genes)
regulation of acetyl-coa biosynthetic process from pyruvate (4 genes)
positive regulation of phosphatidylinositol biosynthetic process (4 genes)
positive regulation of phospholipase activity (8 genes)
positive regulation of calcium ion transport into cytosol (16 genes)
negative regulation of transposition (14 genes)
positive regulation of activation of janus kinase activity (4 genes)
regulation of platelet activation (4 genes)
negative regulation of platelet activation (7 genes)
positive regulation of glycoprotein biosynthetic process (4 genes)
regulation of cell cycle process (8 genes)
regulation of double-strand break repair via homologous recombination (18 genes)
positive regulation of nuclear cell cycle dna replication (4 genes)
positive regulation of platelet activation (6 genes)
regulation of vascular endothelial growth factor production (5 genes)
positive regulation of vascular endothelial growth factor production (24 genes)
mirna metabolic process (11 genes)
mirna catabolic process (7 genes)
regulation of lamellipodium assembly (12 genes)
positive regulation of lamellipodium assembly (23 genes)
regulation of endothelial cell migration (7 genes)
positive regulation of endothelial cell migration (45 genes)
negative regulation of endothelial cell migration (21 genes)
positive regulation of cytoplasmic mrna processing body assembly (5 genes)
posttranscriptional regulation of gene expression (34 genes)
positive regulation of cardiac muscle hypertrophy (25 genes)
negative regulation of cardiac muscle hypertrophy (17 genes)
negative regulation of schwann cell proliferation (5 genes)
positive regulation of gene expression (470 genes)
negative regulation of gene expression (324 genes)
epithelial cell migration (6 genes)
regulation of epithelial cell migration (5 genes)
negative regulation of epithelial cell migration (18 genes)
positive regulation of epithelial cell migration (37 genes)
positive regulation of mitochondrial fusion (4 genes)
negative regulation of mitochondrial fusion (5 genes)
positive regulation of organelle organization (4 genes)
negative regulation of platelet-derived growth factor receptor signaling pathway (10 genes)
cell communication by electrical coupling (5 genes)
negative regulation of muscle cell apoptotic process (11 genes)
cardiac muscle cell apoptotic process (6 genes)
negative regulation of striated muscle cell apoptotic process (4 genes)
positive regulation of cardiac muscle cell apoptotic process (20 genes)
negative regulation of cardiac muscle cell apoptotic process (31 genes)
epithelial structure maintenance (6 genes)
negative regulation of alkaline phosphatase activity (4 genes)
positive regulation of alkaline phosphatase activity (5 genes)
negative regulation of norepinephrine secretion (8 genes)
positive regulation of norepinephrine secretion (6 genes)
regulation of epithelial to mesenchymal transition (13 genes)
positive regulation of epithelial to mesenchymal transition (43 genes)
negative regulation of epithelial to mesenchymal transition (25 genes)
regulation of definitive erythrocyte differentiation (4 genes)
negative regulation of hydrogen peroxide biosynthetic process (4 genes)
positive regulation of transcription via serum response element binding (4 genes)
protein kinase a signaling (10 genes)
regulation of protein kinase a signaling (14 genes)
positive regulation of protein kinase a signaling (11 genes)
macrophage derived foam cell differentiation (5 genes)
positive regulation of macrophage derived foam cell differentiation (11 genes)
negative regulation of macrophage derived foam cell differentiation (10 genes)
negative regulation of plasma membrane long-chain fatty acid transport (4 genes)
regulation of nitric oxide mediated signal transduction (6 genes)
negative regulation of cgmp-mediated signaling (7 genes)
positive regulation of plasminogen activation (6 genes)
negative regulation of plasminogen activation (6 genes)
positive regulation of macrophage chemotaxis (20 genes)
negative regulation of macrophage chemotaxis (7 genes)
fibroblast migration (12 genes)
regulation of fibroblast migration (10 genes)
positive regulation of fibroblast migration (17 genes)
negative regulation of fibroblast migration (8 genes)
positive regulation of sodium ion transport (24 genes)
negative regulation of sodium ion transport (8 genes)
dna double-strand break processing involved in repair via single-strand annealing (5 genes)
regulation of mrna export from nucleus (7 genes)
positive regulation of peptidyl-threonine phosphorylation (29 genes)
negative regulation of peptidyl-threonine phosphorylation (16 genes)
regulation of tumor necrosis factor-mediated signaling pathway (8 genes)
negative regulation of tumor necrosis factor-mediated signaling pathway (14 genes)
regulation of synaptic vesicle priming (8 genes)
positive regulation of cell-substrate adhesion (48 genes)
negative regulation of cell-substrate adhesion (16 genes)
bradykinin catabolic process (7 genes)
regulation of hormone levels (4 genes)
t cell chemotaxis (5 genes)
positive regulation of t cell chemotaxis (14 genes)
regulation of mitochondrion organization (16 genes)
positive regulation of mitochondrion organization (4 genes)
regulation of centrosome duplication (21 genes)
positive regulation of centrosome duplication (5 genes)
negative regulation of centrosome duplication (5 genes)
regulation of glucose transport (6 genes)
positive regulation of glucose transport (10 genes)
negative regulation of glucose transport (5 genes)
regulation of myotube differentiation (6 genes)
positive regulation of myotube differentiation (12 genes)
negative regulation of myotube differentiation (14 genes)
telomere maintenance via telomere lengthening (7 genes)
regulation of protein adp-ribosylation (6 genes)
regulation of keratinocyte proliferation (7 genes)
positive regulation of keratinocyte proliferation (10 genes)
negative regulation of keratinocyte proliferation (15 genes)
positive regulation of circadian sleep/wake cycle, wakefulness (4 genes)
retina layer formation (20 genes)
positive regulation of pathway-restricted smad protein phosphorylation (49 genes)
positive regulation of phospholipase c activity (9 genes)
positive regulation of triglyceride biosynthetic process (17 genes)
positive regulation of receptor biosynthetic process (7 genes)
positive regulation of cholesterol esterification (9 genes)
positive regulation of cholesterol efflux (15 genes)
regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum (14 genes)
regulation of cardiac muscle contraction by regulation of the release of sequestered calcium ion (7 genes)
regulation of cardiac muscle contraction by calcium ion signaling (7 genes)
regulation of lipid storage (4 genes)
positive regulation of lipid storage (11 genes)
positive regulation of cholesterol storage (8 genes)
negative regulation of cholesterol storage (6 genes)
negative regulation of lipid storage (8 genes)
positive regulation of sequestering of triglyceride (7 genes)
negative regulation of sequestering of triglyceride (5 genes)
positive regulation of steroid biosynthetic process (5 genes)
negative regulation of steroid biosynthetic process (6 genes)
negative regulation of triglyceride catabolic process (4 genes)
positive regulation of triglyceride catabolic process (9 genes)
regulation of glucose metabolic process (25 genes)
positive regulation of glucose metabolic process (9 genes)
negative regulation of mitochondrial membrane potential (11 genes)
positive regulation of mitochondrial membrane potential (13 genes)
positive regulation of phosphatase activity (6 genes)
negative regulation of phosphatase activity (50 genes)
negative regulation of macrophage cytokine production (5 genes)
positive regulation of necrotic cell death (7 genes)
regulation of cell death (9 genes)
positive regulation of cell death (58 genes)
negative regulation of transcription by competitive promoter binding (8 genes)
positive regulation of endopeptidase activity (8 genes)
negative regulation of endopeptidase activity (84 genes)
positive regulation of protein processing (16 genes)
negative regulation of protein processing (13 genes)
magnesium ion homeostasis (12 genes)
cellular magnesium ion homeostasis (5 genes)
regulation of phosphate transport (4 genes)
positive regulation of g2/m transition of mitotic cell cycle (27 genes)
negative regulation of g2/m transition of mitotic cell cycle (7 genes)
regulation of neuron projection development (35 genes)
positive regulation of neuron projection development (161 genes)
negative regulation of neuron projection development (81 genes)
regulation of low-density lipoprotein particle clearance (4 genes)
negative regulation of low-density lipoprotein particle clearance (7 genes)
negative regulation of smad protein complex assembly (5 genes)
ubiquitin recycling (4 genes)
free ubiquitin chain polymerization (5 genes)
response to auditory stimulus (5 genes)
programmed cell death (29 genes)
astrocyte development (17 genes)
oligodendrocyte development (21 genes)
microglia development (5 genes)
glial cell proliferation (5 genes)
notochord formation (5 genes)
neural crest formation (6 genes)
mesenchymal cell development (7 genes)
neural crest cell development (22 genes)
neural crest cell differentiation (6 genes)
schwann cell differentiation (9 genes)
regulation of neuron maturation (5 genes)
positive regulation of neuron maturation (6 genes)
negative regulation of neuron maturation (5 genes)
schwann cell development (11 genes)
glutamate secretion (7 genes)
positive regulation of glutamate secretion (12 genes)
negative regulation of glutamate secretion (9 genes)
gamma-aminobutyric acid secretion (4 genes)
negative regulation of gamma-aminobutyric acid secretion (4 genes)
positive regulation of gamma-aminobutyric acid secretion (7 genes)
regulation of dopamine secretion (33 genes)
regulation of norepinephrine secretion (5 genes)
negative regulation of serotonin secretion (5 genes)
phosphatidylinositol 3-kinase signaling (34 genes)
regulation of phosphatidylinositol 3-kinase signaling (9 genes)
negative regulation of phosphatidylinositol 3-kinase signaling (15 genes)
positive regulation of phosphatidylinositol 3-kinase signaling (67 genes)
response to organic cyclic compound (37 genes)
striated muscle tissue development (4 genes)
regulation of somitogenesis (8 genes)
release of sequestered calcium ion into cytosol by sarcoplasmic reticulum (6 genes)
skeletal muscle satellite cell differentiation (4 genes)
regulation of skeletal muscle contraction (4 genes)
response to activity (16 genes)
artery smooth muscle contraction (11 genes)
vein smooth muscle contraction (4 genes)
intestine smooth muscle contraction (4 genes)
vascular smooth muscle contraction (4 genes)
urinary bladder smooth muscle contraction (8 genes)
skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration (5 genes)
myoblast differentiation involved in skeletal muscle regeneration (4 genes)
myoblast migration involved in skeletal muscle regeneration (4 genes)
regulation of skeletal muscle satellite cell proliferation (6 genes)
response to muscle activity (16 genes)
response to muscle activity involved in regulation of muscle adaptation (5 genes)
response to electrical stimulus involved in regulation of muscle adaptation (4 genes)
transition between fast and slow fiber (10 genes)
muscle atrophy (5 genes)
response to denervation involved in regulation of muscle adaptation (8 genes)
cardiac muscle hypertrophy in response to stress (18 genes)
myotube differentiation (17 genes)
myotube cell development (6 genes)
smooth muscle cell migration (8 genes)
regulation of smooth muscle cell migration (6 genes)
positive regulation of smooth muscle cell migration (40 genes)
negative regulation of smooth muscle cell migration (15 genes)
heparan sulfate proteoglycan biosynthetic process (14 genes)
heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process (7 genes)
heparan sulfate proteoglycan biosynthetic process, enzymatic modification (5 genes)
protein transport (605 genes)
dna integration (4 genes)
carbon dioxide transport (8 genes)
oxygen transport (7 genes)
monovalent inorganic cation transport (7 genes)
copper ion import (6 genes)
magnesium ion transport (15 genes)
organic cation transport (5 genes)
ammonium transport (7 genes)
quaternary ammonium group transport (6 genes)
bicarbonate transport (18 genes)
iodide transport (4 genes)
organic anion transport (14 genes)
monocarboxylic acid transport (14 genes)
bile acid and bile salt transport (18 genes)
canalicular bile acid transport (6 genes)
taurine transport (4 genes)
sialic acid transport (5 genes)
citrate transport (5 genes)
urate transport (6 genes)
glucose-6-phosphate transport (5 genes)
glycerol transport (4 genes)
neutral amino acid transport (11 genes)
l-amino acid transport (5 genes)
l-alanine transport (6 genes)
arginine transport (5 genes)
aspartate transport (9 genes)
l-glutamate transport (11 genes)
glycine transport (9 genes)
proline transport (7 genes)
peptide transport (8 genes)
urea transport (6 genes)
aminergic neurotransmitter loading into synaptic vesicle (4 genes)
monoamine transport (8 genes)
atp transport (5 genes)
choline transport (6 genes)
dopamine transport (5 genes)
carnitine transport (5 genes)
folic acid transport (4 genes)
heme transport (6 genes)
cobalamin transport (5 genes)
drug transport (9 genes)
fatty acid transport (15 genes)
long-chain fatty acid transport (12 genes)
phospholipid transport (39 genes)
coenzyme a metabolic process (4 genes)
coenzyme a biosynthetic process (10 genes)
atp synthesis coupled proton transport (17 genes)
electron transport coupled proton transport (4 genes)
atp hydrolysis coupled proton transport (29 genes)
viral process (28 genes)
lipid catabolic process (118 genes)
cellular component organization (9 genes)
detection of bacterium (12 genes)
detection of temperature stimulus (4 genes)
cell growth (9 genes)
vesicle organization (11 genes)
carbohydrate biosynthetic process (9 genes)
wnt signaling pathway (219 genes)
rhodopsin mediated signaling pathway (4 genes)
immunoglobulin mediated immune response (13 genes)
rna metabolic process (11 genes)
rrna catabolic process (20 genes)
trna catabolic process (4 genes)
synaptic vesicle exocytosis (32 genes)
synaptic vesicle targeting (4 genes)
synaptic vesicle docking (13 genes)
synaptic vesicle priming (20 genes)
sterol metabolic process (10 genes)
sterol biosynthetic process (28 genes)
glycoside catabolic process (6 genes)
snrna processing (13 genes)
synaptic vesicle budding from endosome (4 genes)
synaptic vesicle budding from presynaptic endocytic zone membrane (4 genes)
synaptic vesicle maturation (10 genes)
synaptic vesicle uncoating (5 genes)
vesicle-mediated transport (237 genes)
endosomal transport (47 genes)
axon midline choice point recognition (5 genes)
regulation of striated muscle tissue development (5 genes)
iron-sulfur cluster assembly (17 genes)
telomere capping (12 genes)
macroautophagy (31 genes)
positive regulation of macroautophagy (30 genes)
autophagosome membrane docking (5 genes)
regulation of macroautophagy (5 genes)
negative regulation of macroautophagy (12 genes)
attachment of gpi anchor to protein (5 genes)
gap junction assembly (6 genes)
o-glycan processing (7 genes)
phosphorylation (621 genes)
dephosphorylation (112 genes)
female meiosis chromosome segregation (5 genes)
neuron remodeling (13 genes)
morphogenesis of embryonic epithelium (13 genes)
calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules (7 genes)
calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules (38 genes)
dendrite development (40 genes)
posttranscriptional gene silencing (5 genes)
somatic hypermutation of immunoglobulin genes (13 genes)
somatic recombination of immunoglobulin gene segments (7 genes)
gene silencing (7 genes)
cell migration (218 genes)
protein processing (61 genes)
peptide hormone processing (13 genes)
negative regulation of angiogenesis (92 genes)
protein autoprocessing (14 genes)
cytidine to uridine editing (7 genes)
mrna modification (7 genes)
peroxisome membrane biogenesis (4 genes)
protein import into peroxisome matrix (13 genes)
peroxisome fission (9 genes)
protein import into peroxisome matrix, docking (4 genes)
protein ubiquitination (266 genes)
histone modification (6 genes)
histone methylation (25 genes)
histone phosphorylation (10 genes)
histone acetylation (34 genes)
histone ubiquitination (11 genes)
histone deacetylation (34 genes)
histone dephosphorylation (6 genes)
histone deubiquitination (13 genes)
protein deubiquitination (78 genes)
nucleosome positioning (16 genes)
rac protein signal transduction (21 genes)
protein sumoylation (20 genes)
protein desumoylation (10 genes)
poly(a)+ mrna export from nucleus (17 genes)
regulation of transforming growth factor beta receptor signaling pathway (17 genes)
protein import (4 genes)
phospholipid scrambling (8 genes)
drug metabolic process (16 genes)
stem cell division (11 genes)
negative regulation of translation (85 genes)
calcium ion regulated exocytosis (29 genes)
regulation of exocytosis (31 genes)
regulation of calcium ion-dependent exocytosis (11 genes)
peptidyl-diphthamide biosynthetic process from peptidyl-histidine (6 genes)
peptidyl-lysine hydroxylation (7 genes)
peptidyl-glutamic acid carboxylation (4 genes)
n-terminal peptidyl-methionine acetylation (6 genes)
peptidyl-lysine methylation (10 genes)
peptidyl-lysine trimethylation (6 genes)
peptidyl-lysine monomethylation (9 genes)
peptidyl-lysine dimethylation (4 genes)
peptidyl-lysine oxidation (5 genes)
protein polyglutamylation (9 genes)
protein citrullination (5 genes)
peptidyl-serine phosphorylation (154 genes)
peptidyl-threonine phosphorylation (66 genes)
peptidyl-tyrosine phosphorylation (68 genes)
peptidyl-cysteine s-nitrosylation (5 genes)
peptidyl-arginine adp-ribosylation (6 genes)
protein hydroxylation (4 genes)
peptide cross-linking (51 genes)
protein oxidation (4 genes)
peptidyl-methionine modification (4 genes)
peptidyl-arginine methylation (5 genes)
peptidyl-l-cysteine s-palmitoylation (22 genes)
protein o-linked glycosylation via serine (6 genes)
protein o-linked glycosylation via threonine (7 genes)
protein n-linked glycosylation via asparagine (17 genes)
protein-chromophore linkage (11 genes)
protein prenylation (5 genes)
protein farnesylation (4 genes)
protein geranylgeranylation (6 genes)
protein palmitoylation (24 genes)
internal peptidyl-lysine acetylation (5 genes)
peptidyl-lysine acetylation (5 genes)
peptidyl-proline hydroxylation to 4-hydroxy-l-proline (8 genes)
protein c-linked glycosylation via 2'-alpha-mannosyl-l-tryptophan (4 genes)
nitrobenzene metabolic process (4 genes)
modulation by virus of host morphology or physiology (4 genes)
virion assembly (5 genes)
viral release from host cell (4 genes)
viral genome replication (4 genes)
transformation of host cell by virus (4 genes)
regulation of lipid metabolic process (36 genes)
regulation of fatty acid metabolic process (11 genes)
regulation of steroid metabolic process (5 genes)
cytokine-mediated signaling pathway (110 genes)
regulation of metabolic process (7 genes)
transmission of nerve impulse (26 genes)
neuronal action potential propagation (11 genes)
neuronal action potential (35 genes)
regulation of vasoconstriction (17 genes)
proprioception (4 genes)
sensory perception of pain (75 genes)
response to pheromone (104 genes)
methylglyoxal catabolic process to d-lactate via s-lactoyl-glutathione (4 genes)
lactate biosynthetic process from pyruvate (4 genes)
n-acetylneuraminate catabolic process (6 genes)
udp-n-acetylgalactosamine metabolic process (4 genes)
cysteine biosynthetic process (5 genes)
transsulfuration (4 genes)
dolichol metabolic process (4 genes)
pyridine nucleotide biosynthetic process (11 genes)
fatty acid elongation, saturated fatty acid (7 genes)
arachidonic acid metabolic process (27 genes)
leukotriene biosynthetic process (12 genes)
lipoxygenase pathway (9 genes)
epoxygenase p450 pathway (31 genes)
galactose catabolic process (5 genes)
fatty acid oxidation (16 genes)
removal of superoxide radicals (11 genes)
triglyceride biosynthetic process (15 genes)
triglyceride catabolic process (15 genes)
aromatic compound catabolic process (6 genes)
tryptophan catabolic process to kynurenine (5 genes)
l-methionine salvage from methylthioadenosine (5 genes)
taurine metabolic process (7 genes)
oxalate transport (5 genes)
protein metabolic process (13 genes)
gdp-mannose metabolic process (4 genes)
nad metabolic process (7 genes)
ribose phosphate metabolic process (6 genes)
choline metabolic process (4 genes)
calcium-mediated signaling (79 genes)
cellular homeostasis (8 genes)
antimicrobial humoral response (7 genes)
antibacterial humoral response (45 genes)
antifungal humoral response (4 genes)
carboxylic acid metabolic process (16 genes)
peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan (8 genes)
quinolinate biosynthetic process (4 genes)
stem cell population maintenance (69 genes)
cytolysis (19 genes)
l-ascorbic acid metabolic process (4 genes)
l-ascorbic acid biosynthetic process (4 genes)
antigen processing and presentation (32 genes)
antigen processing and presentation of endogenous peptide antigen via mhc class i (5 genes)
antigen processing and presentation of exogenous peptide antigen via mhc class ii (15 genes)
axonal transport of mitochondrion (8 genes)
lipid storage (26 genes)
peptidyl-arginine methylation, to asymmetrical-dimethyl arginine (6 genes)
camp-mediated signaling (24 genes)
cgmp-mediated signaling (26 genes)
modification-dependent protein catabolic process (10 genes)
sexual reproduction (10 genes)
translesion synthesis (11 genes)
hemoglobin metabolic process (6 genes)
spinal cord development (19 genes)
spinal cord dorsal/ventral patterning (9 genes)
dorsal spinal cord development (8 genes)
ventral spinal cord development (9 genes)
spinal cord motor neuron cell fate specification (7 genes)
ventral spinal cord interneuron specification (5 genes)
spinal cord motor neuron differentiation (27 genes)
spinal cord association neuron differentiation (13 genes)
neural tube patterning (6 genes)
cell differentiation in hindbrain (4 genes)
telencephalon development (24 genes)
corpus callosum morphogenesis (8 genes)
dentate gyrus development (18 genes)
subpallium development (5 genes)
cranial nerve development (4 genes)
cerebellum development (37 genes)
optic nerve development (5 genes)
midbrain-hindbrain boundary morphogenesis (4 genes)
trigeminal nerve development (6 genes)
rhombomere 3 development (5 genes)
hindbrain morphogenesis (4 genes)
cerebellum morphogenesis (9 genes)
ventricular system development (21 genes)
facial nerve structural organization (11 genes)
glossopharyngeal nerve morphogenesis (4 genes)
optic nerve morphogenesis (5 genes)
trigeminal nerve structural organization (5 genes)
vestibulocochlear nerve formation (4 genes)
lateral ventricle development (12 genes)
nerve development (14 genes)
third ventricle development (4 genes)
cerebellar purkinje cell layer development (15 genes)
cerebellar cortex development (8 genes)
cerebellar cortex morphogenesis (6 genes)
cerebellar purkinje cell differentiation (13 genes)
cerebellar granule cell differentiation (10 genes)
cochlear nucleus development (4 genes)
striatum development (13 genes)
amygdala development (5 genes)
hippocampus development (59 genes)
olfactory bulb development (23 genes)
glial cell development (6 genes)
branchiomotor neuron axon guidance (6 genes)
thalamus development (12 genes)
cerebral cortex cell migration (13 genes)
cerebral cortex regionalization (9 genes)
forebrain anterior/posterior pattern specification (4 genes)
forebrain dorsal/ventral pattern formation (7 genes)
cerebral cortex radially oriented cell migration (16 genes)
cerebral cortex tangential migration (4 genes)
layer formation in cerebral cortex (14 genes)
cell proliferation in forebrain (15 genes)
cerebral cortex gabaergic interneuron migration (6 genes)
hypothalamus development (7 genes)
pyramidal neuron development (9 genes)
forebrain radial glial cell differentiation (8 genes)
forebrain regionalization (5 genes)
forebrain neuron differentiation (11 genes)
forebrain neuron development (8 genes)
olfactory bulb interneuron differentiation (16 genes)
olfactory bulb interneuron development (5 genes)
cerebral cortex neuron differentiation (15 genes)
commitment of neuronal cell to specific neuron type in forebrain (7 genes)
dorsal/ventral neural tube patterning (19 genes)
regulation of transcription from rna polymerase ii promoter involved in ventral spinal cord interneuron specification (4 genes)
neural tube development (49 genes)
cerebellar granule cell precursor proliferation (6 genes)
smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation (4 genes)
positive regulation of cerebellar granule cell precursor proliferation (5 genes)
radial glia guided migration of purkinje cell (7 genes)
formation of radial glial scaffolds (5 genes)
central nervous system projection neuron axonogenesis (11 genes)
central nervous system neuron differentiation (12 genes)
central nervous system neuron development (16 genes)
central nervous system neuron axonogenesis (10 genes)
corticospinal tract morphogenesis (7 genes)
anterior commissure morphogenesis (6 genes)
telencephalon regionalization (8 genes)
hypothalamus cell differentiation (4 genes)
pituitary gland development (25 genes)
adenohypophysis development (9 genes)
cerebral cortex development (52 genes)
convergent extension involved in neural plate elongation (4 genes)
neurogenesis (63 genes)
central nervous system vasculogenesis (4 genes)
central nervous system myelination (12 genes)
myelination in peripheral nervous system (21 genes)
interkinetic nuclear migration (11 genes)
tangential migration from the subventricular zone to the olfactory bulb (4 genes)
telencephalon cell migration (7 genes)
metencephalon development (4 genes)
corpus callosum development (11 genes)
hair cycle process (7 genes)
regulation of cell-cell adhesion (13 genes)
negative regulation of cell-cell adhesion (20 genes)
positive regulation of cell-cell adhesion (22 genes)
reproductive process (11 genes)
protein maturation by protein folding (6 genes)
regulation of cell morphogenesis (26 genes)
gland morphogenesis (8 genes)
protein to membrane docking (4 genes)
extracellular matrix disassembly (15 genes)
regulation of transmembrane transporter activity (5 genes)
electron transport chain (10 genes)
respiratory electron transport chain (18 genes)
signal transduction by protein phosphorylation (51 genes)
signal transduction involved in regulation of gene expression (23 genes)
neuronal signal transduction (5 genes)
regulation of signaling (4 genes)
signaling (7 genes)
metal ion transport (28 genes)
cellular potassium ion homeostasis (14 genes)
establishment of cell polarity (38 genes)
maintenance of cell polarity (6 genes)
actin filament-based process (4 genes)
cell projection organization (179 genes)
cell projection assembly (21 genes)
lamellipodium assembly (37 genes)
microvillus assembly (10 genes)
microspike assembly (4 genes)
actin cytoskeleton organization (160 genes)
actin filament polymerization (25 genes)
actin filament depolymerization (11 genes)
actin filament fragmentation (4 genes)
actin filament-based movement (18 genes)
muscle filament sliding (7 genes)
vesicle transport along actin filament (5 genes)
regulation of mitotic metaphase/anaphase transition (9 genes)
peptide hormone secretion (5 genes)
insulin secretion (37 genes)
hemopoiesis (82 genes)
lymphocyte differentiation (11 genes)
myeloid cell differentiation (25 genes)
regulation of endocytosis (33 genes)
natural killer cell activation (23 genes)
water homeostasis (4 genes)
regulation of wnt signaling pathway (21 genes)
sphingolipid biosynthetic process (31 genes)
protein import into mitochondrial matrix (18 genes)
cell differentiation (947 genes)
regulation of cell adhesion (39 genes)
pancreatic juice secretion (5 genes)
regulation of proteolysis (19 genes)
protein catabolic process (53 genes)
proteoglycan biosynthetic process (9 genes)
platelet activation (36 genes)
regulation of dna-dependent dna replication initiation (4 genes)
positive regulation of wnt signaling pathway (35 genes)
negative regulation of wnt signaling pathway (51 genes)
neuron differentiation (138 genes)
b cell differentiation (77 genes)
regulation of blood coagulation (11 genes)
positive regulation of blood coagulation (14 genes)
negative regulation of blood coagulation (15 genes)
extracellular matrix organization (133 genes)
collagen fibril organization (49 genes)
glycosaminoglycan metabolic process (6 genes)
chondroitin sulfate metabolic process (4 genes)
chondroitin sulfate biosynthetic process (10 genes)
hyaluronan metabolic process (9 genes)
hyaluronan biosynthetic process (6 genes)
hyaluronan catabolic process (8 genes)
keratinocyte differentiation (77 genes)
t cell differentiation (44 genes)
erythrocyte differentiation (50 genes)
megakaryocyte differentiation (7 genes)
platelet formation (19 genes)
neutrophil differentiation (5 genes)
monocyte differentiation (14 genes)
macrophage differentiation (14 genes)
male sex determination (15 genes)
myofibril assembly (12 genes)
skeletal muscle thin filament assembly (13 genes)
skeletal muscle myosin thick filament assembly (9 genes)
autophagy of peroxisome (5 genes)
growth hormone secretion (6 genes)
lipid glycosylation (13 genes)
chromosome condensation (25 genes)
apoptotic nuclear changes (6 genes)
apoptotic chromosome condensation (5 genes)
maintenance of gastrointestinal epithelium (11 genes)
regulation of ossification (21 genes)
negative regulation of ossification (26 genes)
bone mineralization (39 genes)
intestinal cholesterol absorption (7 genes)
regulation of intestinal cholesterol absorption (5 genes)
cholesterol transport (20 genes)
positive regulation of cell growth (97 genes)
negative regulation of cell growth (127 genes)
poly-n-acetyllactosamine biosynthetic process (10 genes)
osteoclast differentiation (35 genes)
flagellated sperm motility (76 genes)
melanocyte differentiation (20 genes)
transepithelial chloride transport (7 genes)
respiratory tube development (6 genes)
lung development (114 genes)
adrenal gland development (22 genes)
embryonic limb morphogenesis (62 genes)
dna damage response, signal transduction by p53 class mediator (12 genes)
regulation of cell migration (99 genes)
positive regulation of cell migration (232 genes)
negative regulation of cell migration (121 genes)
sperm mitochondrion organization (4 genes)
fructose 1,6-bisphosphate metabolic process (9 genes)
production of sirna involved in rna interference (7 genes)
targeting of mrna for destruction involved in rna interference (4 genes)
sleep (9 genes)
peristalsis (11 genes)
ubiquitin-dependent erad pathway (59 genes)
regulation of complement activation (4 genes)
trna methylation (22 genes)
maturation of ssu-rrna (11 genes)
fatty acid elongation (8 genes)
regulation of bone mineralization (24 genes)
positive regulation of bone mineralization (35 genes)
negative regulation of bone mineralization (15 genes)
bmp signaling pathway (89 genes)
regulation of bmp signaling pathway (9 genes)
positive regulation of transforming growth factor beta receptor signaling pathway (29 genes)
negative regulation of transforming growth factor beta receptor signaling pathway (59 genes)
positive regulation of bmp signaling pathway (37 genes)
negative regulation of bmp signaling pathway (51 genes)
regulation of axon extension (14 genes)
negative regulation of axon extension (26 genes)
intracellular steroid hormone receptor signaling pathway (15 genes)
intracellular estrogen receptor signaling pathway (13 genes)
androgen receptor signaling pathway (23 genes)
intracellular receptor signaling pathway (4 genes)
adult behavior (37 genes)
embryonic genitalia morphogenesis (4 genes)
male genitalia development (19 genes)
female genitalia development (4 genes)
collagen catabolic process (22 genes)
nuclear body organization (4 genes)
cajal body organization (4 genes)
pml body organization (5 genes)
ubiquitin-dependent smad protein catabolic process (6 genes)
neutrophil chemotaxis (78 genes)
leukocyte chemotaxis (25 genes)
regulation of cellular ph (6 genes)
cellular phosphate ion homeostasis (8 genes)
cellular chloride ion homeostasis (9 genes)
cytoskeleton-dependent intracellular transport (12 genes)
germ-line stem cell population maintenance (6 genes)
ovulation (7 genes)
sequestering of triglyceride (4 genes)
regulation of actin filament length (4 genes)
regulation of actin filament polymerization (33 genes)
negative regulation of actin filament depolymerization (6 genes)
positive regulation of actin filament depolymerization (9 genes)
negative regulation of actin filament polymerization (21 genes)
positive regulation of actin filament polymerization (50 genes)
prostate gland development (7 genes)
granulocyte differentiation (11 genes)
negative regulation of granulocyte differentiation (9 genes)
positive regulation of granulocyte differentiation (7 genes)
epithelial cell differentiation (46 genes)
regulation of epithelial cell differentiation (9 genes)
negative regulation of epithelial cell differentiation (12 genes)
positive regulation of epithelial cell differentiation (8 genes)
cortical cytoskeleton organization (8 genes)
cortical actin cytoskeleton organization (24 genes)
thyroid gland development (27 genes)
mammary gland development (62 genes)
negative regulation of myeloid dendritic cell activation (4 genes)
regulation of b cell proliferation (5 genes)
negative regulation of b cell proliferation (19 genes)
positive regulation of b cell proliferation (44 genes)
forebrain development (78 genes)
midbrain development (38 genes)
hindbrain development (22 genes)
notochord development (14 genes)
olfactory placode formation (4 genes)
paranodal junction assembly (7 genes)
otic vesicle formation (7 genes)
midbrain-hindbrain boundary development (5 genes)
regulation of vascular endothelial growth factor receptor signaling pathway (5 genes)
negative regulation of vascular endothelial growth factor receptor signaling pathway (13 genes)
positive regulation of vascular endothelial growth factor receptor signaling pathway (14 genes)
establishment or maintenance of actin cytoskeleton polarity (14 genes)
establishment or maintenance of microtubule cytoskeleton polarity (4 genes)
astral microtubule organization (10 genes)
endoplasmic reticulum unfolded protein response (42 genes)
retrograde protein transport, er to cytosol (16 genes)
response to caffeine (9 genes)
pancreas development (32 genes)
exocrine pancreas development (9 genes)
endocrine pancreas development (26 genes)
microtubule organizing center organization (10 genes)
actomyosin structure organization (26 genes)
gene silencing by rna (55 genes)
primary mirna processing (8 genes)
pre-mirna processing (14 genes)
regulation of histone methylation (4 genes)
positive regulation of histone methylation (9 genes)
regulation of histone deacetylation (5 genes)
positive regulation of histone deacetylation (15 genes)
hair follicle morphogenesis (32 genes)
embryonic camera-type eye development (13 genes)
post-embryonic camera-type eye development (5 genes)
deadenylation-independent decapping of nuclear-transcribed mrna (4 genes)
stress-activated protein kinase signaling cascade (53 genes)
animal organ regeneration (14 genes)
neuron projection regeneration (5 genes)
axon regeneration (13 genes)
regulation of microtubule polymerization or depolymerization (14 genes)
regulation of microtubule polymerization (9 genes)
negative regulation of microtubule polymerization (14 genes)
positive regulation of microtubule polymerization (26 genes)
positive regulation of microtubule depolymerization (4 genes)
trna pseudouridine synthesis (5 genes)
cytoplasmic microtubule organization (46 genes)
rna 3'-end processing (8 genes)
mrna 3'-end processing (4 genes)
regulation of axon diameter (6 genes)
anaphase-promoting complex-dependent catabolic process (13 genes)
scf-dependent proteasomal ubiquitin-dependent protein catabolic process (41 genes)
rrna methylation (9 genes)
neuron projection development (161 genes)
biomineral tissue development (34 genes)
auditory behavior (6 genes)
positive regulation of pseudopodium assembly (10 genes)
regulation of guanylate cyclase activity (4 genes)
positive regulation of guanylate cyclase activity (7 genes)
retinal ganglion cell axon guidance (19 genes)
membrane protein intracellular domain proteolysis (6 genes)
t cell costimulation (23 genes)
b cell costimulation (4 genes)
replication fork processing (25 genes)
negative regulation of protein complex assembly (20 genes)
positive regulation of protein complex assembly (24 genes)
positive regulation of vesicle fusion (9 genes)
negative regulation of cell projection organization (4 genes)
positive regulation of cell projection organization (8 genes)
positive regulation of prostaglandin biosynthetic process (6 genes)
regulation of protein ubiquitination (18 genes)
negative regulation of protein ubiquitination (49 genes)
positive regulation of protein ubiquitination (69 genes)
positive regulation of protein modification process (4 genes)
keratinization (31 genes)
positive regulation of mrna 3'-end processing (7 genes)
nuclear envelope reassembly (7 genes)
chromatin assembly (9 genes)
protein complex localization (4 genes)
pericentric heterochromatin assembly (4 genes)
ruffle organization (12 genes)
actin cytoskeleton reorganization (59 genes)
positive regulation of exit from mitosis (9 genes)
brain-derived neurotrophic factor receptor signaling pathway (4 genes)
mitotic g1 dna damage checkpoint (9 genes)
intra-s dna damage checkpoint (14 genes)
membrane raft organization (7 genes)
hemidesmosome assembly (5 genes)
activation of phospholipase d activity (7 genes)
positive regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity (4 genes)
cell-substrate adhesion (19 genes)
wybutosine biosynthetic process (4 genes)
positive regulation of fever generation (7 genes)
receptor internalization (40 genes)
synaptic vesicle fusion to presynaptic active zone membrane (14 genes)
regulation of synaptic vesicle fusion to presynaptic active zone membrane (7 genes)
zymogen activation (26 genes)
plasminogen activation (12 genes)
killing of cells of other organism (13 genes)
regulation of myelination (25 genes)
negative regulation of myelination (8 genes)
positive regulation of myelination (18 genes)
regulation of protein stability (73 genes)
protein destabilization (45 genes)
heat generation (4 genes)
positive regulation of heat generation (4 genes)
lipopolysaccharide-mediated signaling pathway (32 genes)
negative regulation of lipopolysaccharide-mediated signaling pathway (6 genes)
positive regulation of lipopolysaccharide-mediated signaling pathway (8 genes)
response to nutrient levels (35 genes)
cellular response to extracellular stimulus (20 genes)
cellular response to nutrient levels (10 genes)
cellular response to nutrient (4 genes)
protection from non-homologous end joining at telomere (7 genes)
telomeric 3' overhang formation (4 genes)
negative regulation of synaptic plasticity (6 genes)
positive regulation of synaptic plasticity (12 genes)
tor signaling (17 genes)
negative regulation of chromatin silencing (14 genes)
positive regulation of chromatin silencing (4 genes)
regulation of protein autophosphorylation (10 genes)
negative regulation of protein autophosphorylation (13 genes)
positive regulation of protein autophosphorylation (18 genes)
locomotion involved in locomotory behavior (8 genes)
regulation of fatty acid beta-oxidation (5 genes)
negative regulation of fatty acid beta-oxidation (5 genes)
positive regulation of fatty acid beta-oxidation (10 genes)
regulation of tor signaling (19 genes)
negative regulation of tor signaling (31 genes)
positive regulation of tor signaling (31 genes)
regulation of arf protein signal transduction (16 genes)
isg15-protein conjugation (5 genes)
positive regulation of insulin secretion (52 genes)
response to magnesium ion (6 genes)
mitochondrial dna metabolic process (4 genes)
cardiolipin biosynthetic process (8 genes)
ciliary basal body organization (4 genes)
bleb assembly (11 genes)
positive regulation of endodeoxyribonuclease activity (4 genes)
negative regulation of nf-kappab transcription factor activity (75 genes)
negative regulation of protein binding (75 genes)
positive regulation of protein binding (84 genes)
response to food (15 genes)
regulation of response to food (5 genes)
regulation of appetite (5 genes)
negative regulation of appetite (10 genes)
activation of protein kinase activity (76 genes)
activation of protein kinase b activity (20 genes)
regulation of telomere maintenance (12 genes)
negative regulation of telomere maintenance (5 genes)
positive regulation of telomere maintenance (11 genes)
regulation of telomere maintenance via telomerase (5 genes)
negative regulation of telomere maintenance via telomerase (20 genes)
positive regulation of telomere maintenance via telomerase (33 genes)
positive regulation of synaptic transmission, cholinergic (7 genes)
regulation of synaptic transmission, dopaminergic (4 genes)
positive regulation of synaptic transmission, dopaminergic (4 genes)
negative regulation of synaptic transmission, dopaminergic (4 genes)
regulation of synaptic transmission, gabaergic (12 genes)
negative regulation of synaptic transmission, gabaergic (8 genes)
positive regulation of synaptic transmission, gabaergic (16 genes)
negative regulation of actin filament bundle assembly (6 genes)
positive regulation of actin filament bundle assembly (14 genes)
activation of store-operated calcium channel activity (5 genes)
secretory granule localization (4 genes)
methylation (170 genes)
imp salvage (4 genes)
regulation of cellular protein metabolic process (6 genes)
negative regulation of cellular protein metabolic process (6 genes)
positive regulation of cellular protein metabolic process (17 genes)
positive regulation of protein polymerization (5 genes)
luteinizing hormone secretion (4 genes)
central nervous system myelin maintenance (4 genes)
peripheral nervous system myelin maintenance (8 genes)
myelin assembly (7 genes)
peripheral nervous system myelin formation (4 genes)
positive regulation of prostaglandin secretion (9 genes)
molybdopterin cofactor biosynthetic process (4 genes)
regulation of chondrocyte differentiation (9 genes)
negative regulation of chondrocyte differentiation (19 genes)
positive regulation of chondrocyte differentiation (21 genes)
negative regulation of aldosterone biosynthetic process (4 genes)
regulation of hormone metabolic process (5 genes)
response to estradiol (29 genes)
oxygen homeostasis (5 genes)
intracellular cholesterol transport (10 genes)
negative regulation of lipid transport (5 genes)
regulation of cholesterol transport (5 genes)
positive regulation of cholesterol transport (4 genes)
regulation of intracellular transport (4 genes)
positive regulation of intracellular transport (6 genes)
melanosome localization (6 genes)
melanosome transport (17 genes)
negative regulation of transporter activity (6 genes)
positive regulation of transporter activity (4 genes)
positive regulation of ion transmembrane transporter activity (9 genes)
positive regulation of sodium:proton antiporter activity (4 genes)
lysosome localization (21 genes)
regulation of proteasomal ubiquitin-dependent protein catabolic process (11 genes)
negative regulation of proteasomal ubiquitin-dependent protein catabolic process (30 genes)
positive regulation of proteasomal ubiquitin-dependent protein catabolic process (72 genes)
melanosome organization (12 genes)
endocytic recycling (32 genes)
negative regulation of protein oligomerization (5 genes)
positive regulation of protein oligomerization (15 genes)
negative regulation of protein homooligomerization (9 genes)
positive regulation of protein homooligomerization (11 genes)
regulation of cytokinesis (38 genes)
negative regulation of cytokinesis (6 genes)
positive regulation of cytokinesis (30 genes)
endoplasmic reticulum calcium ion homeostasis (15 genes)
negative regulation of endoplasmic reticulum calcium ion concentration (9 genes)
negative regulation of type i interferon production (9 genes)
positive regulation of type i interferon production (18 genes)
rab protein signal transduction (66 genes)
rap protein signal transduction (9 genes)
cdc42 protein signal transduction (6 genes)
regulation of cdc42 protein signal transduction (6 genes)
response to peptidoglycan (9 genes)
response to muramyl dipeptide (15 genes)
response to lipopolysaccharide (106 genes)
detection of lipopolysaccharide (5 genes)
developmental process (13 genes)
maintenance of protein location in cell (6 genes)
dna duplex unwinding (18 genes)
endosome transport via multivesicular body sorting pathway (15 genes)
endosome to lysosome transport via multivesicular body sorting pathway (4 genes)
late endosome to vacuole transport via multivesicular body sorting pathway (4 genes)
negative regulation of phosphoprotein phosphatase activity (15 genes)
positive regulation of phosphoprotein phosphatase activity (14 genes)
somite rostral/caudal axis specification (13 genes)
response to retinoic acid (41 genes)
regulation of microvillus length (7 genes)
regulation of microvillus assembly (7 genes)
mitochondrial translation (33 genes)
response to progesterone (14 genes)
interferon-gamma production (13 genes)
interleukin-1 alpha production (4 genes)
interleukin-1 beta production (11 genes)
interleukin-10 production (4 genes)
interleukin-17 production (4 genes)
interleukin-2 production (5 genes)
interleukin-6 production (6 genes)
tumor necrosis factor production (4 genes)
regulation of chemokine production (5 genes)
regulation of interferon-beta production (4 genes)
regulation of interferon-gamma production (5 genes)
regulation of interleukin-6 production (6 genes)
regulation of tumor necrosis factor production (9 genes)
negative regulation of interferon-alpha production (4 genes)
negative regulation of interferon-beta production (13 genes)
negative regulation of interferon-gamma production (28 genes)
negative regulation of interleukin-1 beta production (13 genes)
negative regulation of interleukin-10 production (16 genes)
negative regulation of interleukin-12 production (15 genes)
negative regulation of interleukin-17 production (13 genes)
negative regulation of interleukin-2 production (18 genes)
negative regulation of interleukin-4 production (6 genes)
negative regulation of interleukin-5 production (4 genes)
negative regulation of interleukin-6 production (33 genes)
negative regulation of interleukin-8 production (5 genes)
negative regulation of tumor necrosis factor production (50 genes)
positive regulation of chemokine production (23 genes)
positive regulation of granulocyte macrophage colony-stimulating factor production (9 genes)
positive regulation of interferon-alpha production (13 genes)
positive regulation of interferon-beta production (27 genes)
positive regulation of interferon-gamma production (55 genes)
positive regulation of interleukin-1 beta production (10 genes)
positive regulation of interleukin-1 production (5 genes)
positive regulation of interleukin-10 production (26 genes)
positive regulation of interleukin-12 production (25 genes)
positive regulation of interleukin-13 production (9 genes)
positive regulation of interleukin-17 production (12 genes)
positive regulation of interleukin-18 production (6 genes)
positive regulation of interleukin-2 production (13 genes)
positive regulation of interleukin-23 production (5 genes)
positive regulation of interleukin-4 production (22 genes)
positive regulation of interleukin-5 production (7 genes)
positive regulation of interleukin-6 production (62 genes)
positive regulation of interleukin-8 production (27 genes)
positive regulation of tumor necrosis factor production (63 genes)
negative regulation of atpase activity (12 genes)
positive regulation of atpase activity (25 genes)
bile acid secretion (6 genes)
regulation of dna-templated transcription, elongation (6 genes)
positive regulation of dna-templated transcription, elongation (10 genes)
ribosome disassembly (7 genes)
negative regulation of creb transcription factor activity (6 genes)
positive regulation of creb transcription factor activity (16 genes)
receptor catabolic process (12 genes)
lacrimal gland development (7 genes)
negative regulation of epinephrine secretion (7 genes)
negative regulation of natural killer cell activation (4 genes)
positive regulation of natural killer cell activation (12 genes)
positive regulation of natural killer cell proliferation (6 genes)
positive regulation of natural killer cell differentiation (12 genes)
negative regulation of natural killer cell differentiation involved in immune response (4 genes)
glomerulus development (9 genes)
glomerular basement membrane development (8 genes)
positive regulation of cellular ph reduction (4 genes)
response to insulin (47 genes)
cellular response to insulin stimulus (86 genes)
cellular response to hormone stimulus (29 genes)
regulation of stress-activated mapk cascade (9 genes)
negative regulation of stress-activated mapk cascade (8 genes)
positive regulation of stress-activated mapk cascade (22 genes)
negative regulation of dna endoreduplication (7 genes)
regulation of establishment or maintenance of cell polarity (4 genes)
regulation of protein localization (70 genes)
regulation of microtubule-based process (13 genes)
negative regulation of viral transcription (12 genes)
regulation of transforming growth factor beta2 production (4 genes)
negative regulation of transforming growth factor beta1 production (4 genes)
positive regulation of transforming growth factor beta1 production (5 genes)
circadian regulation of gene expression (62 genes)
activin receptor signaling pathway (15 genes)
regulation of activin receptor signaling pathway (5 genes)
negative regulation of activin receptor signaling pathway (10 genes)
positive regulation of activin receptor signaling pathway (8 genes)
negative regulation of superoxide anion generation (4 genes)
positive regulation of superoxide anion generation (20 genes)
srebp signaling pathway (8 genes)
secretion by cell (10 genes)
positive regulation of mononuclear cell proliferation (4 genes)
regulation of actin cytoskeleton organization (67 genes)
inositol trisphosphate metabolic process (5 genes)
inositol phosphate biosynthetic process (11 genes)
inositol trisphosphate biosynthetic process (4 genes)
positive regulation of inositol trisphosphate biosynthetic process (6 genes)
collagen metabolic process (15 genes)
collagen biosynthetic process (9 genes)
negative regulation of collagen biosynthetic process (14 genes)
positive regulation of collagen biosynthetic process (31 genes)
positive regulation of transcription elongation from rna polymerase ii promoter (23 genes)
regulation of actin filament-based process (5 genes)
mitochondrial respiratory chain complex i assembly (43 genes)
negative regulation of mast cell activation (6 genes)
positive regulation of mast cell activation (7 genes)
tetrapyrrole biosynthetic process (5 genes)
negative regulation of mast cell apoptotic process (4 genes)
negative regulation of neutrophil apoptotic process (4 genes)
positive regulation of neutrophil apoptotic process (5 genes)
negative regulation of myeloid cell apoptotic process (6 genes)
regulation of chromosome organization (4 genes)
t cell differentiation in thymus (37 genes)
regulation of t cell differentiation in thymus (6 genes)
negative regulation of t cell differentiation in thymus (6 genes)
negative regulation of immature t cell proliferation in thymus (5 genes)
positive regulation of t cell differentiation in thymus (12 genes)
positive regulation of immature t cell proliferation in thymus (4 genes)
mitochondrial respiratory chain complex assembly (4 genes)
negative regulation of rna splicing (7 genes)
positive regulation of rna splicing (17 genes)
positive regulation of histone phosphorylation (9 genes)
negative regulation of glucokinase activity (5 genes)
positive regulation of glucokinase activity (6 genes)
regulation of peptidyl-serine phosphorylation (6 genes)
negative regulation of peptidyl-serine phosphorylation (26 genes)
positive regulation of peptidyl-serine phosphorylation (91 genes)
positive regulation of peptidyl-serine phosphorylation of stat protein (28 genes)
negative regulation of intracellular steroid hormone receptor signaling pathway (4 genes)
regulation of intracellular estrogen receptor signaling pathway (11 genes)
negative regulation of intracellular estrogen receptor signaling pathway (13 genes)
positive regulation of intracellular estrogen receptor signaling pathway (13 genes)
v(d)j recombination (9 genes)
immunoglobulin v(d)j recombination (5 genes)
t cell receptor v(d)j recombination (5 genes)
regulation of intracellular protein transport (6 genes)
regulation of protein import into nucleus, translocation (4 genes)
negative regulation of protein import into nucleus, translocation (4 genes)
positive regulation of protein import into nucleus, translocation (18 genes)
histone h3-k9 demethylation (14 genes)
calcineurin-nfat signaling cascade (12 genes)
response to hydroperoxide (10 genes)
response to atp (18 genes)
tumor necrosis factor-mediated signaling pathway (28 genes)
leptin-mediated signaling pathway (10 genes)
adiponectin-activated signaling pathway (5 genes)
dsrna transport (5 genes)
negative regulation of protein sumoylation (9 genes)
positive regulation of protein sumoylation (14 genes)
nuclear dna replication (6 genes)
regulation of nuclear cell cycle dna replication (5 genes)
cell proliferation in midbrain (6 genes)
response to vitamin d (5 genes)
t-tubule organization (5 genes)
secretion of lysosomal enzymes (7 genes)
mitotic dna replication checkpoint (10 genes)
cerebrospinal fluid secretion (4 genes)
leydig cell differentiation (12 genes)
cholesterol efflux (25 genes)
secretory granule organization (4 genes)
protein localization to organelle (23 genes)
putrescine biosynthetic process from ornithine (4 genes)
galactose catabolic process via udp-galactose (5 genes)
floor plate development (4 genes)
histone h2a ubiquitination (5 genes)
histone h2b ubiquitination (9 genes)
fatty acid beta-oxidation using acyl-coa dehydrogenase (10 genes)
fatty acid beta-oxidation using acyl-coa oxidase (4 genes)
cellular response to stress (6 genes)
multicellular organismal response to stress (11 genes)
regulation of water loss via skin (4 genes)
dorsal/ventral axon guidance (4 genes)
anterior/posterior axon guidance (5 genes)
transferrin transport (10 genes)
response to testosterone (11 genes)
mammary gland epithelial cell proliferation (10 genes)
regulation of mammary gland epithelial cell proliferation (5 genes)
negative regulation of mammary gland epithelial cell proliferation (6 genes)
positive regulation of mammary gland epithelial cell proliferation (9 genes)
negative regulation of dopamine secretion (5 genes)
positive regulation of dopamine secretion (11 genes)
positive regulation of catecholamine secretion (5 genes)
mitochondrial proton-transporting atp synthase complex assembly (4 genes)
mitochondrial respiratory chain complex iv assembly (18 genes)
membrane protein proteolysis (10 genes)
integrin activation (9 genes)
positive regulation of integrin activation (7 genes)
cell adhesion mediated by integrin (23 genes)
regulation of cell adhesion mediated by integrin (10 genes)
negative regulation of cell adhesion mediated by integrin (5 genes)
positive regulation of cell adhesion mediated by integrin (14 genes)
regulation of cell-cell adhesion mediated by integrin (4 genes)
positive regulation of cell-cell adhesion mediated by integrin (6 genes)
negative regulation of kinase activity (13 genes)
positive regulation of kinase activity (14 genes)
nucleotide-excision repair, dna incision (6 genes)
negative regulation of luteinizing hormone secretion (5 genes)
positive regulation of luteinizing hormone secretion (4 genes)
negative regulation of osteoblast proliferation (9 genes)
positive regulation of osteoblast proliferation (14 genes)
phospholipid efflux (11 genes)
response to glucagon (5 genes)
positive regulation of nad(p)h oxidase activity (5 genes)
cytoplasmic mrna processing body assembly (14 genes)
response to lipid (18 genes)
endosomal vesicle fusion (8 genes)
stress granule assembly (15 genes)
protein localization to golgi apparatus (20 genes)
cenp-a containing nucleosome assembly (8 genes)
maintenance of mitotic sister chromatid cohesion (4 genes)
response to cytokine (63 genes)
erythrocyte homeostasis (12 genes)
regulation of tissue remodeling (4 genes)
homotypic cell-cell adhesion (9 genes)
negative regulation of homotypic cell-cell adhesion (4 genes)
heterotypic cell-cell adhesion (19 genes)
negative regulation of heterotypic cell-cell adhesion (12 genes)
positive regulation of heterotypic cell-cell adhesion (13 genes)
regulation of toll-like receptor signaling pathway (4 genes)
negative regulation of toll-like receptor signaling pathway (17 genes)
positive regulation of toll-like receptor signaling pathway (5 genes)
toll-like receptor 2 signaling pathway (5 genes)
negative regulation of toll-like receptor 2 signaling pathway (5 genes)
positive regulation of toll-like receptor 2 signaling pathway (7 genes)
toll-like receptor 3 signaling pathway (9 genes)
positive regulation of toll-like receptor 3 signaling pathway (7 genes)
toll-like receptor 4 signaling pathway (11 genes)
negative regulation of toll-like receptor 4 signaling pathway (8 genes)
positive regulation of toll-like receptor 4 signaling pathway (10 genes)
toll-like receptor 7 signaling pathway (4 genes)
positive regulation of toll-like receptor 7 signaling pathway (4 genes)
toll-like receptor 9 signaling pathway (6 genes)
positive regulation of toll-like receptor 9 signaling pathway (6 genes)
positive regulation of maintenance of mitotic sister chromatid cohesion (5 genes)
triglyceride transport (4 genes)
cellular response to amino acid starvation (41 genes)
amyloid-beta formation (6 genes)
protein hexamerization (10 genes)
ion transmembrane transport (74 genes)
trna thio-modification (4 genes)
negative regulation of transcription elongation from rna polymerase ii promoter (13 genes)
negative regulation of gtpase activity (31 genes)
arp2/3 complex-mediated actin nucleation (16 genes)
negative regulation of arp2/3 complex-mediated actin nucleation (6 genes)
cell junction assembly (7 genes)
adherens junction organization (29 genes)
adherens junction assembly (7 genes)
adherens junction maintenance (5 genes)
response to type i interferon (7 genes)
response to interferon-gamma (25 genes)
negative regulation of glial cell apoptotic process (7 genes)
very-low-density lipoprotein particle remodeling (7 genes)
low-density lipoprotein particle remodeling (9 genes)
high-density lipoprotein particle remodeling (11 genes)
very-low-density lipoprotein particle assembly (7 genes)
high-density lipoprotein particle assembly (8 genes)
chylomicron remnant clearance (4 genes)
low-density lipoprotein particle clearance (12 genes)
high-density lipoprotein particle clearance (6 genes)
lipid particle organization (16 genes)
regulation of smooth muscle cell apoptotic process (4 genes)
negative regulation of smooth muscle cell apoptotic process (9 genes)
positive regulation of smooth muscle cell apoptotic process (9 genes)
protein localization to cell surface (23 genes)
response to fluid shear stress (6 genes)
nuclear-transcribed mrna catabolic process, exonucleolytic, 3'-5' (9 genes)
cholesterol esterification (5 genes)
substrate adhesion-dependent cell spreading (43 genes)
microtubule anchoring (16 genes)
microtubule anchoring at centrosome (10 genes)
snrna 3'-end processing (8 genes)
u4 snrna 3'-end processing (8 genes)
protein localization to phagophore assembly site (13 genes)
early endosome to golgi transport (8 genes)
late endosome to golgi transport (4 genes)
protein localization to kinetochore (11 genes)
protein localization to chromosome (6 genes)
protein localization to nucleus (41 genes)
mitochondrial respiratory chain complex iii assembly (9 genes)
mitochondrial respiratory chain complex ii assembly (4 genes)
pirna metabolic process (15 genes)
cellular response to oxidative stress (67 genes)
cellular response to heat (59 genes)
response to tumor necrosis factor (23 genes)
cellular protein localization (56 genes)
cellular response to reactive oxygen species (33 genes)
cellular response to unfolded protein (20 genes)
cellular macromolecular complex assembly (21 genes)
fatty acid elongation, monounsaturated fatty acid (7 genes)
fatty acid elongation, polyunsaturated fatty acid (7 genes)
cellular protein complex localization (11 genes)
phosphatidylcholine catabolic process (4 genes)
cellular response to uv (49 genes)
retinoic acid catabolic process (4 genes)
response to prostaglandin e (5 genes)
histone h3-k4 demethylation (7 genes)
regulation of ion transmembrane transport (140 genes)
positive regulation of ion transmembrane transport (5 genes)
histone h4-k20 methylation (4 genes)
histone h4-k20 trimethylation (6 genes)
histone lysine methylation (13 genes)
histone arginine methylation (9 genes)
protein folding in endoplasmic reticulum (10 genes)
response to endoplasmic reticulum stress (70 genes)
mitochondrial protein processing (5 genes)
peptidyl-lysine deacetylation (7 genes)
somatic stem cell population maintenance (45 genes)
regulation of rac protein signal transduction (6 genes)
negative regulation of rac protein signal transduction (5 genes)
positive regulation of rac protein signal transduction (4 genes)
regulation of rho protein signal transduction (77 genes)
negative regulation of rho protein signal transduction (22 genes)
positive regulation of rho protein signal transduction (32 genes)
sperm-egg recognition (5 genes)
embryonic heart tube development (24 genes)
regulation of histone acetylation (7 genes)
positive regulation of histone acetylation (19 genes)
negative regulation of histone acetylation (11 genes)
axoneme assembly (19 genes)
sirna loading onto risc involved in rna interference (4 genes)
establishment or maintenance of apical/basal cell polarity (4 genes)
establishment of apical/basal cell polarity (4 genes)
sperm chromatin condensation (6 genes)
spermatogenesis, exchange of chromosomal proteins (10 genes)
response to nicotine (26 genes)
behavioral response to nicotine (9 genes)
operant conditioning (5 genes)
limb morphogenesis (28 genes)
embryonic forelimb morphogenesis (36 genes)
embryonic hindlimb morphogenesis (34 genes)
forelimb morphogenesis (14 genes)
hindlimb morphogenesis (16 genes)
tube formation (8 genes)
embryonic hemopoiesis (21 genes)
post-embryonic hemopoiesis (6 genes)
social behavior (51 genes)
gene silencing by mirna (63 genes)
production of mirnas involved in gene silencing by mirna (18 genes)
ectopic germ cell programmed cell death (10 genes)
ionotropic glutamate receptor signaling pathway (14 genes)
tube morphogenesis (9 genes)
synaptic transmission, glutamatergic (34 genes)
multicellular organism growth (125 genes)
organ growth (15 genes)
protein o-linked mannosylation (9 genes)
mirna mediated inhibition of translation (16 genes)
mrna cleavage involved in gene silencing by mirna (5 genes)
mirna loading onto risc involved in gene silencing by mirna (7 genes)
brain segmentation (4 genes)
negative regulation of dephosphorylation (4 genes)
positive regulation of protein dephosphorylation (23 genes)
negative regulation of protein dephosphorylation (12 genes)
wound healing, spreading of epidermal cells (11 genes)
hair cell differentiation (5 genes)
hippo signaling (20 genes)
regulation of hippo signaling (4 genes)
negative regulation of hippo signaling (7 genes)
peptidyl-tyrosine dephosphorylation (26 genes)
long-chain fatty-acyl-coa metabolic process (5 genes)
cellular triglyceride homeostasis (7 genes)
peroxisome proliferator activated receptor signaling pathway (7 genes)
negative regulation of peroxisome proliferator activated receptor signaling pathway (4 genes)
positive regulation of peroxisome proliferator activated receptor signaling pathway (8 genes)
protein localization to microtubule (11 genes)
roundabout signaling pathway (9 genes)
protein localization to synapse (17 genes)
autocrine signaling (5 genes)
extracellular matrix-cell signaling (4 genes)
phosphate ion transmembrane transport (13 genes)
response to interferon-alpha (12 genes)
response to interferon-beta (11 genes)
cellular response to interferon-alpha (12 genes)
cellular response to interferon-beta (53 genes)
cargo loading into vesicle (4 genes)
determination of pancreatic left/right asymmetry (5 genes)
positive regulation of vascular wound healing (5 genes)
positive regulation of notch signaling pathway involved in heart induction (5 genes)
gastric emptying (4 genes)
snare complex assembly (11 genes)
negative regulation of myosin-light-chain-phosphatase activity (6 genes)
histone h2a monoubiquitination (12 genes)
protein k29-linked ubiquitination (5 genes)
monoubiquitinated protein deubiquitination (6 genes)
monoubiquitinated histone h2a deubiquitination (4 genes)
protein k29-linked deubiquitination (5 genes)
proline transmembrane transport (5 genes)
regulation of snare complex assembly (5 genes)
positive regulation of interferon-beta secretion (5 genes)
intracellular signal transduction (424 genes)
negative regulation of chromatin binding (9 genes)
positive regulation of chromatin binding (14 genes)
non-canonical wnt signaling pathway (24 genes)
sequestering of bmp in extracellular matrix (4 genes)
sequestering of tgfbeta in extracellular matrix (4 genes)
calcium-mediated signaling using intracellular calcium source (19 genes)
protein side chain deglutamylation (5 genes)
histone h2b conserved c-terminal lysine deubiquitination (4 genes)
stress granule disassembly (5 genes)
intrahepatic bile duct development (4 genes)
ceramide transport (8 genes)
bone mineralization involved in bone maturation (5 genes)
maintenance of permeability of blood-brain barrier (5 genes)
exploration behavior (17 genes)
locomotory exploration behavior (15 genes)
endosome to melanosome transport (10 genes)
interleukin-18-mediated signaling pathway (5 genes)
trif-dependent toll-like receptor signaling pathway (4 genes)
cellular response to drug (52 genes)
mitochondrial protein catabolic process (5 genes)
hematopoietic stem cell migration (5 genes)
intraciliary anterograde transport (5 genes)
intraciliary retrograde transport (12 genes)
sodium ion transmembrane transport (35 genes)
common myeloid progenitor cell proliferation (12 genes)
cellular response to hepatocyte growth factor stimulus (14 genes)
intraciliary transport involved in cilium assembly (7 genes)
t-helper 1 cell cytokine production (5 genes)
t-helper 2 cell cytokine production (4 genes)
regulation of lysosomal lumen ph (4 genes)
b cell chemotaxis (5 genes)
endothelial cell chemotaxis (10 genes)
insulin secretion involved in cellular response to glucose stimulus (8 genes)
positive regulation of insulin secretion involved in cellular response to glucose stimulus (38 genes)
platelet-derived growth factor receptor-beta signaling pathway (4 genes)
positive regulation of mitochondrial membrane permeability (4 genes)
ureter maturation (4 genes)
positive regulation of urine volume (13 genes)
negative regulation of urine volume (9 genes)
negative regulation of renal sodium excretion (9 genes)
positive regulation of renal sodium excretion (14 genes)
photoreceptor cell outer segment organization (11 genes)
megakaryocyte development (18 genes)
glial cell-derived neurotrophic factor receptor signaling pathway (5 genes)
cellular response to potassium ion (7 genes)
protein k11-linked deubiquitination (10 genes)
nail development (6 genes)
plasma membrane lactate transport (4 genes)
embryonic nail plate morphogenesis (4 genes)
amacrine cell differentiation (7 genes)
response to immobilization stress (5 genes)
aorta development (27 genes)
aorta morphogenesis (16 genes)
ascending aorta morphogenesis (4 genes)
dorsal aorta morphogenesis (8 genes)
skeletal muscle cell differentiation (55 genes)
cellular response to vascular endothelial growth factor stimulus (26 genes)
positive regulation of gluconeogenesis by positive regulation of transcription from rna polymerase ii promoter (5 genes)
peptidyl-threonine dephosphorylation (13 genes)
aggrephagy (5 genes)
cellular response to trichostatin a (4 genes)
endodermal cell differentiation (11 genes)
chondrocyte proliferation (10 genes)
tendon development (4 genes)
response to muscle stretch (15 genes)
detection of muscle stretch (4 genes)
tetrahydrofolate interconversion (11 genes)
positive regulation of transcription from rna polymerase ii promoter in response to stress (5 genes)
cellular response to macrophage colony-stimulating factor stimulus (6 genes)
protein localization to endosome (7 genes)
cellular response to interleukin-3 (4 genes)
osteoclast development (5 genes)
fucosylation (11 genes)
protein o-linked fucosylation (7 genes)
cleavage furrow formation (7 genes)
positive regulation of transcription from rna polymerase ii promoter in response to oxidative stress (4 genes)
phosphatidylinositol-3-phosphate biosynthetic process (8 genes)
cellular response to platelet-derived growth factor stimulus (18 genes)
histone h3-k9 trimethylation (6 genes)
outer dynein arm assembly (17 genes)
inner dynein arm assembly (14 genes)
peptidyl-serine autophosphorylation (8 genes)
cellular response to decreased oxygen levels (25 genes)
cellular response to increased oxygen levels (7 genes)
interstrand cross-link repair (23 genes)
atrioventricular canal development (7 genes)
dendritic cell migration (5 genes)
post-anal tail morphogenesis (25 genes)
psychomotor behavior (4 genes)
platelet maturation (4 genes)
histone h2a-k119 monoubiquitination (9 genes)
sodium ion export across plasma membrane (11 genes)
histone citrullination (5 genes)
maintenance of lens transparency (5 genes)
mitochondrial calcium uptake (10 genes)
synaptic vesicle recycling (7 genes)
ventral trunk neural crest cell migration (4 genes)
erad pathway (12 genes)
dopaminergic neuron axon guidance (4 genes)
serotonergic neuron axon guidance (4 genes)
paracrine signaling (7 genes)
opioid receptor signaling pathway (4 genes)
netrin-activated signaling pathway (7 genes)
reelin-mediated signaling pathway (4 genes)
apolipoprotein a-i-mediated signaling pathway (4 genes)
non-canonical wnt signaling pathway via jnk cascade (5 genes)
positive regulation of endothelial cell chemotaxis by vegf-activated vascular endothelial growth factor receptor signaling pathway (6 genes)
nik/nf-kappab signaling (6 genes)
collagen-activated tyrosine kinase receptor signaling pathway (12 genes)
collagen-activated signaling pathway (4 genes)
p38mapk cascade (8 genes)
peptidyl-tyrosine autophosphorylation (34 genes)
vascular endothelial growth factor signaling pathway (11 genes)
nodal signaling pathway (9 genes)
fc-gamma receptor signaling pathway (4 genes)
negative regulation of appetite by leptin-mediated signaling pathway (4 genes)
interleukin-3-mediated signaling pathway (4 genes)
thrombopoietin-mediated signaling pathway (4 genes)
oncostatin-m-mediated signaling pathway (4 genes)
angiotensin-activated signaling pathway (8 genes)
nerve growth factor signaling pathway (11 genes)
torc1 signaling (10 genes)
torc2 signaling (6 genes)
negative regulation of mda-5 signaling pathway (4 genes)
negative regulation of rig-i signaling pathway (7 genes)
negative stranded viral rna replication (4 genes)
viral rna genome replication (8 genes)
viral budding via host escrt complex (9 genes)
regulation of growth (79 genes)
positive regulation of growth rate (4 genes)
locomotion (11 genes)
regulation of locomotion (8 genes)
negative regulation of locomotion (4 genes)
regulation of multicellular organism growth (36 genes)
negative regulation of multicellular organism growth (17 genes)
embryonic cleavage (8 genes)
positive regulation of multicellular organism growth (43 genes)
positive regulation of embryonic development (10 genes)
regulation of meiotic nuclear division (8 genes)
regulation of gene expression, epigenetic (18 genes)
regulation of fibroblast growth factor receptor signaling pathway (6 genes)
negative regulation of fibroblast growth factor receptor signaling pathway (16 genes)
polar body extrusion after meiotic divisions (6 genes)
protein refolding (19 genes)
regulation of cytokine biosynthetic process (5 genes)
epithelial fluid transport (7 genes)
olfactory behavior (7 genes)
regulation of dopamine metabolic process (10 genes)
regulation of epidermal growth factor receptor signaling pathway (8 genes)
negative regulation of epidermal growth factor receptor signaling pathway (15 genes)
wound healing (74 genes)
gliogenesis (7 genes)
intraciliary transport (29 genes)
cell migration involved in gastrulation (11 genes)
t-helper 1 type immune response (8 genes)
t-helper cell differentiation (4 genes)
t cell proliferation (25 genes)
b cell proliferation (42 genes)
positive regulation of t cell proliferation (65 genes)
positive regulation of activated t cell proliferation (27 genes)
cytokine metabolic process (6 genes)
t cell activation (37 genes)
b cell activation (30 genes)
macrophage activation (8 genes)
endothelial cell activation (7 genes)
neutrophil activation (10 genes)
regulation of cell proliferation (180 genes)
regulation of t cell proliferation (11 genes)
negative regulation of t cell proliferation (49 genes)
neurotransmitter catabolic process (11 genes)
neurotransmitter biosynthetic process (8 genes)
meiotic dna double-strand break formation (6 genes)
retrograde transport, endosome to golgi (69 genes)
strand invasion (4 genes)
cellular response to glucose starvation (38 genes)
lipoprotein metabolic process (18 genes)
lipoprotein biosynthetic process (6 genes)
lipoprotein catabolic process (4 genes)
heme catabolic process (4 genes)
heme metabolic process (7 genes)
regulation of protein catabolic process (18 genes)
negative regulation of protein catabolic process (38 genes)
xenobiotic catabolic process (8 genes)
response to cocaine (21 genes)
tissue regeneration (8 genes)
ribosome biogenesis (91 genes)
ribosome assembly (5 genes)
mature ribosome assembly (6 genes)
peptidyl-aspartic acid hydroxylation (4 genes)
natural killer cell mediated cytotoxicity (21 genes)
protection from natural killer cell mediated cytotoxicity (6 genes)
susceptibility to natural killer cell mediated cytotoxicity (15 genes)
ribosomal large subunit biogenesis (28 genes)
ribosomal small subunit biogenesis (16 genes)
error-prone translesion synthesis (5 genes)
purine nucleoside metabolic process (5 genes)
vocal learning (8 genes)
regulation of fatty acid biosynthetic process (4 genes)
regulation of protein import into nucleus (6 genes)
positive regulation of protein import into nucleus (39 genes)
negative regulation of protein import into nucleus (14 genes)
vasoconstriction (18 genes)
vasodilation (24 genes)
regulation of phosphorylation (17 genes)
negative regulation of phosphorylation (18 genes)
positive regulation of phosphorylation (35 genes)
vitamin d metabolic process (4 genes)
regulation of membrane potential (126 genes)
thyroid hormone metabolic process (12 genes)
cristae formation (14 genes)
norepinephrine metabolic process (10 genes)
dopamine biosynthetic process (12 genes)
dopamine metabolic process (16 genes)
dopamine catabolic process (7 genes)
norepinephrine biosynthetic process (9 genes)
catecholamine biosynthetic process (6 genes)
serotonin metabolic process (9 genes)
melanin biosynthetic process (14 genes)
hormone metabolic process (16 genes)
hormone biosynthetic process (8 genes)
hormone catabolic process (4 genes)
progesterone metabolic process (10 genes)
photoreceptor cell development (5 genes)
eye photoreceptor cell development (19 genes)
ear morphogenesis (6 genes)
inner ear morphogenesis (72 genes)
outer ear morphogenesis (9 genes)
middle ear morphogenesis (24 genes)
odontogenesis of dentin-containing tooth (56 genes)
odontogenesis (23 genes)
regulation of odontogenesis (4 genes)
positive regulation of odontogenesis (5 genes)
regulation of odontogenesis of dentin-containing tooth (8 genes)
mechanoreceptor differentiation (7 genes)
inner ear auditory receptor cell differentiation (16 genes)
response to drug (144 genes)
serine phosphorylation of stat protein (5 genes)
regulation of tyrosine phosphorylation of stat protein (7 genes)
positive regulation of tyrosine phosphorylation of stat protein (55 genes)
negative regulation of tyrosine phosphorylation of stat protein (13 genes)
positive regulation of tumor necrosis factor biosynthetic process (13 genes)
negative regulation of tumor necrosis factor biosynthetic process (5 genes)
hyperosmotic salinity response (5 genes)
hemoglobin biosynthetic process (7 genes)
response to hydrogen peroxide (39 genes)
neuron maturation (10 genes)
myelination (66 genes)
superoxide anion generation (11 genes)
retinol metabolic process (18 genes)
retinoic acid metabolic process (12 genes)
retinal metabolic process (13 genes)
antigen processing and presentation of exogenous peptide antigen via mhc class i (8 genes)
homeostatic process (6 genes)
glucose homeostasis (132 genes)
response to starvation (18 genes)
fear response (8 genes)
cholesterol homeostasis (83 genes)
hair cycle (11 genes)
regulation of hair cycle (7 genes)
catagen (4 genes)
anagen (12 genes)
negative regulation of mesodermal cell fate specification (4 genes)
muscle cell differentiation (16 genes)
ovulation cycle (6 genes)
follicle-stimulating hormone signaling pathway (6 genes)
maternal behavior (17 genes)
sperm ejaculation (6 genes)
fibrinolysis (15 genes)
embryonic digit morphogenesis (68 genes)
exogenous drug catabolic process (53 genes)
defense response to bacterium (146 genes)
hydrogen peroxide metabolic process (8 genes)
hydrogen peroxide catabolic process (25 genes)
regulation of circadian rhythm (51 genes)
positive regulation of circadian rhythm (12 genes)
negative regulation of circadian rhythm (8 genes)
eating behavior (38 genes)
drinking behavior (15 genes)
long-chain fatty acid catabolic process (5 genes)
long-chain fatty acid biosynthetic process (6 genes)
very long-chain fatty acid catabolic process (6 genes)
very long-chain fatty acid biosynthetic process (13 genes)
dna damage response, detection of dna damage (7 genes)
signal transduction in response to dna damage (11 genes)
intrinsic apoptotic signaling pathway in response to dna damage by p53 class mediator (31 genes)
atp synthesis coupled electron transport (6 genes)
mitochondrial atp synthesis coupled electron transport (11 genes)
mitochondrial atp synthesis coupled proton transport (4 genes)
mrna transcription by rna polymerase ii (19 genes)
snrna transcription by rna polymerase ii (8 genes)
snrna transcription by rna polymerase iii (6 genes)
defense response to protozoan (32 genes)
pyruvate biosynthetic process (6 genes)
9-cis-retinoic acid biosynthetic process (7 genes)
glucocorticoid receptor signaling pathway (4 genes)
lipoprotein transport (14 genes)
activation of janus kinase activity (7 genes)
regulation of apoptotic process (204 genes)
amyloid precursor protein metabolic process (9 genes)
negative regulation of amyloid precursor protein biosynthetic process (6 genes)
amyloid precursor protein catabolic process (6 genes)
sequestering of actin monomers (12 genes)
cytoplasmic sequestering of transcription factor (9 genes)
negative regulation of golgi to plasma membrane protein transport (4 genes)
positive regulation of golgi to plasma membrane protein transport (5 genes)
golgi to plasma membrane protein transport (25 genes)
chordate embryonic development (6 genes)
camera-type eye development (72 genes)
myeloid dendritic cell differentiation (20 genes)
t cell homeostasis (31 genes)
regulation of macrophage activation (11 genes)
negative regulation of macrophage activation (6 genes)
positive regulation of macrophage activation (21 genes)
trna aminoacylation (13 genes)
peptide biosynthetic process (4 genes)
atp-dependent chromatin remodeling (15 genes)
dna methylation involved in embryo development (5 genes)
dna methylation involved in gamete generation (19 genes)
positive regulation of apoptotic process (341 genes)
negative regulation of apoptotic process (588 genes)
regulation of programmed cell death (4 genes)
positive regulation of programmed cell death (10 genes)
negative regulation of programmed cell death (15 genes)
penile erection (5 genes)
positive regulation of catalytic activity (40 genes)
negative regulation of catalytic activity (27 genes)
regulation of gtpase activity (80 genes)
replication fork arrest (4 genes)
receptor clustering (31 genes)
negative regulation of vascular permeability (14 genes)
positive regulation of vascular permeability (14 genes)
regulation of i-kappab kinase/nf-kappab signaling (19 genes)
positive regulation of i-kappab kinase/nf-kappab signaling (104 genes)
negative regulation of i-kappab kinase/nf-kappab signaling (44 genes)
surfactant homeostasis (15 genes)
dna replication, removal of rna primer (4 genes)
stress fiber assembly (15 genes)
induction of bacterial agglutination (4 genes)
entrainment of circadian clock by photoperiod (23 genes)
negative regulation of cysteine-type endopeptidase activity involved in apoptotic process (66 genes)
proteasome-mediated ubiquitin-dependent protein catabolic process (167 genes)
ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway (12 genes)
peptide catabolic process (14 genes)
response to amino acid (12 genes)
myelin maintenance (4 genes)
positive regulation of protein complex disassembly (6 genes)
telomere maintenance in response to dna damage (4 genes)
proteasome assembly (12 genes)
erythrocyte maturation (14 genes)
sodium-independent organic anion transport (18 genes)
regulation of protein complex assembly (12 genes)
regulation of potassium ion transport (7 genes)
negative regulation of potassium ion transport (14 genes)
positive regulation of potassium ion transport (13 genes)
positive regulation of ion transport (7 genes)
negative regulation of ion transport (5 genes)
anoikis (10 genes)
apoptotic cell clearance (18 genes)
response to morphine (17 genes)
positive regulation of cysteine-type endopeptidase activity involved in apoptotic process (60 genes)
regulation of cysteine-type endopeptidase activity involved in apoptotic process (12 genes)
apical junction assembly (8 genes)
mast cell degranulation (10 genes)
regulation of mast cell degranulation (8 genes)
negative regulation of mast cell degranulation (6 genes)
positive regulation of mast cell degranulation (16 genes)
neutrophil degranulation (4 genes)
positive regulation of neutrophil degranulation (4 genes)
natural killer cell degranulation (5 genes)
protein transport to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway (4 genes)
response to exogenous dsrna (43 genes)
enucleate erythrocyte differentiation (6 genes)
cd4-positive, alpha-beta t cell differentiation (5 genes)
positive regulation of cd4-positive, alpha-beta t cell differentiation (6 genes)
cd8-positive, alpha-beta t cell differentiation (6 genes)
positive regulation of cd8-positive, alpha-beta t cell differentiation (4 genes)
positive regulation of dna binding (34 genes)
negative regulation of dna binding (33 genes)
regulation of protein binding (24 genes)
steroid hormone mediated signaling pathway (7 genes)
skeletal muscle tissue regeneration (19 genes)
regulation of map kinase activity (10 genes)
positive regulation of map kinase activity (55 genes)
negative regulation of map kinase activity (41 genes)
regulation of mapk cascade (65 genes)
negative regulation of mapk cascade (28 genes)
positive regulation of mapk cascade (115 genes)
positive regulation of skeletal muscle tissue regeneration (4 genes)
negative regulation of dna binding transcription factor activity (76 genes)
response to peptide hormone (44 genes)
regulation of cellular respiration (7 genes)
regulation of atpase activity (6 genes)
pigmentation (46 genes)
regulation of rna splicing (34 genes)
histone exchange (8 genes)
regulation of rna stability (4 genes)
regulation of mrna stability (32 genes)
malate-aspartate shuttle (5 genes)
protein kinase b signaling (41 genes)
regulation of protein homodimerization activity (7 genes)
regulation of protein heterodimerization activity (10 genes)
mitochondrial dna repair (6 genes)
positive regulation of jun kinase activity (41 genes)
negative regulation of jun kinase activity (14 genes)
regulation of dna damage response, signal transduction by p53 class mediator (5 genes)
positive regulation of dna damage response, signal transduction by p53 class mediator (11 genes)
negative regulation of dna damage response, signal transduction by p53 class mediator (16 genes)
regulation of neuron apoptotic process (31 genes)
negative regulation of neuron apoptotic process (165 genes)
positive regulation of neuron apoptotic process (75 genes)
blood vessel endothelial cell migration (9 genes)
regulation of blood vessel endothelial cell migration (5 genes)
positive regulation of blood vessel endothelial cell migration (40 genes)
negative regulation of blood vessel endothelial cell migration (17 genes)
endothelial cell migration (26 genes)
positive regulation of gtpase activity (158 genes)
regulation of kinase activity (6 genes)
regulation of lipid kinase activity (5 genes)
regulation of phosphatidylinositol 3-kinase activity (9 genes)
positive regulation of phosphatidylinositol 3-kinase activity (27 genes)
negative regulation of phosphatidylinositol 3-kinase activity (4 genes)
regulation of insulin-like growth factor receptor signaling pathway (4 genes)
positive regulation of insulin-like growth factor receptor signaling pathway (11 genes)
negative regulation of insulin-like growth factor receptor signaling pathway (9 genes)
maintenance of dna repeat elements (5 genes)
regulation of respiratory gaseous exchange (8 genes)
ear development (15 genes)
nose development (7 genes)
tongue development (12 genes)
tongue morphogenesis (4 genes)
skin development (54 genes)
skin morphogenesis (11 genes)
astrocyte cell migration (6 genes)
keratinocyte proliferation (13 genes)
regulation of transcription from rna polymerase ii promoter in response to oxidative stress (7 genes)
regulation of dna-templated transcription in response to stress (5 genes)
cortical microtubule organization (4 genes)
response to estrogen (37 genes)
rna polyadenylation (5 genes)
dicarboxylic acid metabolic process (4 genes)
linoleic acid metabolic process (10 genes)
engulfment of apoptotic cell (9 genes)
mitochondrial fragmentation involved in apoptotic process (11 genes)
recognition of apoptotic cell (5 genes)
regulation of phosphoprotein phosphatase activity (14 genes)
post-translational protein modification (5 genes)
reverse cholesterol transport (13 genes)
cell dedifferentiation (6 genes)
negative regulation by host of viral transcription (10 genes)
positive regulation by host of viral transcription (16 genes)
exonucleolytic nuclear-transcribed mrna catabolic process involved in deadenylation-dependent decay (9 genes)
ossification involved in bone maturation (8 genes)
regulation of camp-mediated signaling (6 genes)
positive regulation of camp-mediated signaling (32 genes)
negative regulation of camp-mediated signaling (19 genes)
histone h3 acetylation (44 genes)
histone h4 acetylation (30 genes)
histone h2a acetylation (16 genes)
histone h4-k5 acetylation (15 genes)
histone h4-k8 acetylation (15 genes)
histone h4-k12 acetylation (8 genes)
histone h4-k16 acetylation (14 genes)
histone h4-r3 methylation (4 genes)
histone h3-s10 phosphorylation (5 genes)
hypermethylation of cpg island (5 genes)
regulation of dna methylation (17 genes)
regulation of anion transport (5 genes)
growth of symbiont in host (4 genes)
negative regulation of growth of symbiont in host (20 genes)
cellular metabolic process (12 genes)
cellular lipid catabolic process (6 genes)
cellular lipid metabolic process (33 genes)
cellular protein catabolic process (16 genes)
cellular protein metabolic process (9 genes)
cell wall disruption in other organism (6 genes)
protein k27-linked ubiquitination (4 genes)
wound healing, spreading of cells (14 genes)
cellular response to leptin stimulus (7 genes)
response to leptin (9 genes)
cell-cell adhesion mediated by cadherin (26 genes)
canonical wnt signaling pathway involved in negative regulation of apoptotic process (4 genes)
type b pancreatic cell proliferation (12 genes)
cellular response to fibroblast growth factor stimulus (17 genes)
macropinocytosis (7 genes)
negative regulation of protein kinase activity by regulation of protein phosphorylation (8 genes)
adhesion of symbiont to host (9 genes)
motile cilium assembly (30 genes)
regulation of mitochondrial mrna stability (5 genes)
long-chain fatty acid import (9 genes)
daunorubicin metabolic process (8 genes)
doxorubicin metabolic process (8 genes)
cell cycle phase transition (7 genes)
mitotic cell cycle phase transition (5 genes)
cilium organization (17 genes)
positive regulation by host of viral release from host cell (6 genes)
positive regulation by host of viral process (8 genes)
g-quadruplex dna unwinding (6 genes)
negative regulation by host of viral genome replication (7 genes)
positive regulation by host of viral genome replication (8 genes)
cell quiescence (5 genes)
estrous cycle (7 genes)
dna deamination (4 genes)
actin nucleation (11 genes)
negative regulation of nitric oxide biosynthetic process (17 genes)
early endosome to late endosome transport (21 genes)
protein import into mitochondrial inner membrane (9 genes)
protein import into mitochondrial outer membrane (4 genes)
protein import into peroxisome membrane (6 genes)
protein targeting to er (10 genes)
protein retention in golgi apparatus (5 genes)
regulated exocytosis (17 genes)
transcytosis (18 genes)
positive thymic t cell selection (14 genes)
negative thymic t cell selection (15 genes)
thymic t cell selection (5 genes)
t-helper 1 cell differentiation (5 genes)
t-helper 2 cell differentiation (4 genes)
regulatory t cell differentiation (4 genes)
regulation of viral genome replication (8 genes)
positive regulation of viral genome replication (25 genes)
negative regulation of viral genome replication (41 genes)
positive regulation of interferon-gamma biosynthetic process (12 genes)
positive regulation of chemokine biosynthetic process (9 genes)
positive regulation of interleukin-12 biosynthetic process (8 genes)
negative regulation of interleukin-2 biosynthetic process (5 genes)
positive regulation of interleukin-2 biosynthetic process (13 genes)
innate immune response (351 genes)
regulation of innate immune response (11 genes)
positive regulation of innate immune response (28 genes)
intermediate filament cytoskeleton organization (20 genes)
intermediate filament organization (21 genes)
intermediate filament bundle assembly (7 genes)
protein neddylation (14 genes)
cellular extravasation (4 genes)
regulation of bone resorption (8 genes)
meiotic chromosome segregation (7 genes)
meiotic telomere clustering (11 genes)
homologous chromosome segregation (7 genes)
clustering of voltage-gated sodium channels (6 genes)
cell fate commitment (72 genes)
apical protein localization (11 genes)
establishment of protein localization (46 genes)
regulation of circadian sleep/wake cycle, sleep (6 genes)
isotype switching (18 genes)
regulation of isotype switching (4 genes)
establishment or maintenance of epithelial cell apical/basal polarity (18 genes)
establishment of epithelial cell apical/basal polarity (14 genes)
maintenance of epithelial cell apical/basal polarity (5 genes)
neurotransmitter receptor metabolic process (4 genes)
sarcomere organization (38 genes)
cell-cell junction organization (25 genes)
cell-cell junction maintenance (8 genes)
mrna cis splicing, via spliceosome (23 genes)
late endosome to vacuole transport (10 genes)
phospholipid translocation (26 genes)
cellular respiration (20 genes)
positive regulation of mhc class i biosynthetic process (6 genes)
negative regulation of mhc class ii biosynthetic process (4 genes)
positive regulation of mhc class ii biosynthetic process (8 genes)
type i interferon biosynthetic process (5 genes)
positive regulation of interferon-alpha biosynthetic process (4 genes)
positive regulation of interferon-beta biosynthetic process (7 genes)
negative regulation of interleukin-6 biosynthetic process (8 genes)
positive regulation of interleukin-6 biosynthetic process (10 genes)
negative regulation of interleukin-8 biosynthetic process (4 genes)
positive regulation of interleukin-8 biosynthetic process (9 genes)
regulation of nitric oxide biosynthetic process (4 genes)
positive regulation of nitric oxide biosynthetic process (47 genes)
fat cell differentiation (81 genes)
myoblast differentiation (18 genes)
endothelial cell differentiation (7 genes)
bone resorption (23 genes)
cell redox homeostasis (63 genes)
response to ethanol (47 genes)
locomotor rhythm (15 genes)
photoreceptor cell maintenance (41 genes)
regulation of cholesterol biosynthetic process (4 genes)
negative regulation of cholesterol biosynthetic process (5 genes)
positive regulation of cholesterol biosynthetic process (7 genes)
mast cell activation (10 genes)
regulation of b cell differentiation (11 genes)
negative regulation of b cell differentiation (4 genes)
positive regulation of b cell differentiation (18 genes)
regulation of t cell differentiation (13 genes)
negative regulation of t cell differentiation (6 genes)
positive regulation of t cell differentiation (23 genes)
positive regulation of gamma-delta t cell differentiation (6 genes)
regulation of regulatory t cell differentiation (10 genes)
negative regulation of regulatory t cell differentiation (5 genes)
positive regulation of regulatory t cell differentiation (12 genes)
regulation of cell differentiation (32 genes)
negative regulation of cell differentiation (49 genes)
positive regulation of cell differentiation (52 genes)
regulation of fat cell differentiation (21 genes)
negative regulation of fat cell differentiation (52 genes)
positive regulation of fat cell differentiation (51 genes)
regulation of endothelial cell differentiation (5 genes)
negative regulation of endothelial cell differentiation (8 genes)
positive regulation of endothelial cell differentiation (12 genes)
positive regulation of epidermal cell differentiation (7 genes)
regulation of inner ear auditory receptor cell differentiation (7 genes)
regulation of keratinocyte differentiation (9 genes)
negative regulation of keratinocyte differentiation (9 genes)
positive regulation of keratinocyte differentiation (14 genes)
regulation of lymphocyte differentiation (4 genes)
positive regulation of lymphocyte differentiation (5 genes)
positive regulation of t-helper 1 cell differentiation (8 genes)
negative regulation of t-helper 2 cell differentiation (6 genes)
positive regulation of t-helper 2 cell differentiation (7 genes)
positive regulation of melanocyte differentiation (5 genes)
regulation of myeloid cell differentiation (5 genes)
negative regulation of myeloid cell differentiation (22 genes)
positive regulation of myeloid cell differentiation (6 genes)
regulation of erythrocyte differentiation (9 genes)
negative regulation of erythrocyte differentiation (14 genes)
positive regulation of erythrocyte differentiation (24 genes)
negative regulation of macrophage differentiation (6 genes)
positive regulation of macrophage differentiation (13 genes)
regulation of megakaryocyte differentiation (6 genes)
negative regulation of megakaryocyte differentiation (17 genes)
positive regulation of megakaryocyte differentiation (8 genes)
negative regulation of monocyte differentiation (7 genes)
positive regulation of monocyte differentiation (8 genes)
regulation of myoblast differentiation (6 genes)
negative regulation of myoblast differentiation (30 genes)
positive regulation of myoblast differentiation (28 genes)
regulation of neuron differentiation (33 genes)
negative regulation of neuron differentiation (78 genes)
positive regulation of neuron differentiation (109 genes)
regulation of osteoblast differentiation (17 genes)
negative regulation of osteoblast differentiation (64 genes)
positive regulation of osteoblast differentiation (72 genes)
regulation of osteoclast differentiation (17 genes)
negative regulation of osteoclast differentiation (25 genes)
positive regulation of osteoclast differentiation (26 genes)
regulation of epidermis development (4 genes)
positive regulation of epidermis development (4 genes)
positive regulation of glial cell differentiation (9 genes)
negative regulation of fatty acid biosynthetic process (9 genes)
negative regulation of glycogen biosynthetic process (5 genes)
negative regulation of gluconeogenesis (16 genes)
positive regulation of gluconeogenesis (12 genes)
positive regulation of fatty acid biosynthetic process (14 genes)
positive regulation of cilium assembly (15 genes)
positive regulation of glycogen biosynthetic process (17 genes)
positive regulation of translation (69 genes)
respiratory burst (9 genes)
positive regulation of protein catabolic process (74 genes)
negative regulation of cyclin-dependent protein serine/threonine kinase activity (25 genes)
positive regulation of cyclin-dependent protein serine/threonine kinase activity (21 genes)
positive regulation of dna repair (33 genes)
positive regulation of dna replication (33 genes)
positive regulation of epidermal growth factor-activated receptor activity (10 genes)
positive regulation of epidermal growth factor receptor signaling pathway (16 genes)
positive regulation of fibroblast growth factor receptor signaling pathway (7 genes)
negative regulation of g-protein coupled receptor protein signaling pathway (13 genes)
positive regulation of g-protein coupled receptor protein signaling pathway (16 genes)
negative regulation of notch signaling pathway (34 genes)
positive regulation of notch signaling pathway (36 genes)
negative regulation of action potential (5 genes)
positive regulation of action potential (11 genes)
regulation of adenylate cyclase activity (5 genes)
positive regulation of adenylate cyclase activity (10 genes)
regulation of angiogenesis (28 genes)
positive regulation of angiogenesis (147 genes)
positive regulation of axon extension (42 genes)
negative regulation of blood pressure (35 genes)
positive regulation of blood pressure (32 genes)
positive regulation of ossification (22 genes)
negative regulation of bone resorption (15 genes)
positive regulation of bone resorption (28 genes)
positive regulation of cell adhesion (61 genes)
negative regulation of cell cycle (42 genes)
positive regulation of cell cycle (59 genes)
negative regulation of cell size (13 genes)
positive regulation of cell size (16 genes)
negative regulation of cell volume (7 genes)
negative regulation of endocytosis (12 genes)
positive regulation of endocytosis (29 genes)
negative regulation of gene expression, epigenetic (9 genes)
positive regulation of gene expression, epigenetic (5 genes)
negative regulation of glycolytic process (13 genes)
positive regulation of glycolytic process (16 genes)
negative regulation of heart contraction (11 genes)
positive regulation of heart contraction (8 genes)
negative regulation of innate immune response (14 genes)
positive regulation of isotype switching (11 genes)
positive regulation of lipid metabolic process (4 genes)
negative regulation of meiotic nuclear division (7 genes)
positive regulation of meiotic nuclear division (5 genes)
negative regulation of membrane potential (4 genes)
positive regulation of membrane potential (7 genes)
negative regulation of mitotic nuclear division (10 genes)
positive regulation of mitotic nuclear division (33 genes)
positive regulation of mitotic metaphase/anaphase transition (7 genes)
negative regulation of striated muscle tissue development (10 genes)
ph reduction (4 genes)
regulation of protein kinase activity (19 genes)
positive regulation of protein kinase activity (62 genes)
negative regulation of proteolysis (27 genes)
positive regulation of proteolysis (23 genes)
negative regulation of single stranded viral rna replication via double stranded dna intermediate (4 genes)
positive regulation of sister chromatid cohesion (4 genes)
negative regulation of smoothened signaling pathway (26 genes)
positive regulation of smoothened signaling pathway (37 genes)
negative regulation of transcription, dna-templated (536 genes)
positive regulation of transcription, dna-templated (621 genes)
positive regulation of rna polymerase ii transcriptional preinitiation complex assembly (13 genes)
negative regulation of vasoconstriction (10 genes)
positive regulation of vasoconstriction (45 genes)
negative regulation of dna recombination (19 genes)
negative regulation of complement activation (5 genes)
positive regulation of cytolysis (5 genes)
negative regulation of exocytosis (4 genes)
positive regulation of exocytosis (28 genes)
positive regulation of female receptivity (6 genes)
negative regulation of growth (10 genes)
positive regulation of growth (9 genes)
negative regulation of mitotic cell cycle (36 genes)
positive regulation of mitotic cell cycle (40 genes)
positive regulation of transcription by rna polymerase i (13 genes)
positive regulation of transcription by rna polymerase ii (1150 genes)
positive regulation of transcription by rna polymerase iii (12 genes)
negative regulation of translational initiation (13 genes)
positive regulation of translational initiation (13 genes)
negative regulation of mitotic recombination (4 genes)
negative regulation of natural killer cell mediated cytotoxicity (8 genes)
positive regulation of natural killer cell mediated cytotoxicity (22 genes)
negative regulation of calcium ion-dependent exocytosis (10 genes)
positive regulation of calcium ion-dependent exocytosis (16 genes)
negative regulation of dopamine metabolic process (4 genes)
positive regulation of dopamine metabolic process (4 genes)
negative regulation of smooth muscle contraction (8 genes)
positive regulation of smooth muscle contraction (21 genes)
negative regulation of striated muscle contraction (4 genes)
regulation of embryonic development (18 genes)
negative regulation of activated t cell proliferation (11 genes)
positive regulation of circadian sleep/wake cycle, non-rem sleep (9 genes)
amp metabolic process (6 genes)
atp metabolic process (43 genes)
gtp metabolic process (15 genes)
imp metabolic process (4 genes)
datp metabolic process (4 genes)
cgmp metabolic process (4 genes)
cgmp catabolic process (5 genes)
adenosine metabolic process (9 genes)
glycerol-3-phosphate biosynthetic process (4 genes)
regulation of fatty acid oxidation (4 genes)
positive regulation of fatty acid oxidation (8 genes)
negative regulation of fatty acid oxidation (7 genes)
glucose import (8 genes)
regulation of glucose import (8 genes)
negative regulation of glucose import (20 genes)
positive regulation of glucose import (36 genes)
regulation of jnk cascade (20 genes)
negative regulation of jnk cascade (27 genes)
positive regulation of jnk cascade (86 genes)
diacylglycerol metabolic process (14 genes)
diacylglycerol catabolic process (4 genes)
urate metabolic process (10 genes)
regulation of jak-stat cascade (11 genes)
negative regulation of jak-stat cascade (16 genes)
positive regulation of jak-stat cascade (45 genes)
creatinine metabolic process (4 genes)
acylglycerol catabolic process (5 genes)
phosphatidylcholine metabolic process (12 genes)
glycerophospholipid biosynthetic process (5 genes)
glycerophospholipid catabolic process (12 genes)
ether lipid metabolic process (5 genes)
glycerolipid metabolic process (13 genes)
phosphatidylinositol metabolic process (22 genes)
s-adenosylhomocysteine metabolic process (6 genes)
s-adenosylmethionine metabolic process (7 genes)
sphingosine biosynthetic process (9 genes)
ceramide biosynthetic process (27 genes)
negative regulation of photoreceptor cell differentiation (6 genes)
saliva secretion (6 genes)
development of secondary female sexual characteristics (4 genes)
development of primary male sexual characteristics (4 genes)
retinal rod cell development (12 genes)
retinal cone cell development (17 genes)
regulation of ras protein signal transduction (6 genes)
positive regulation of ras protein signal transduction (23 genes)
negative regulation of ras protein signal transduction (25 genes)
regulation of viral entry into host cell (4 genes)
negative regulation of viral entry into host cell (22 genes)
positive regulation of viral entry into host cell (9 genes)
regulation of centriole replication (7 genes)
negative regulation of centriole replication (6 genes)
regulation of mitotic centrosome separation (7 genes)
positive regulation of mitotic centrosome separation (4 genes)
regulation of centrosome cycle (6 genes)
drug export (5 genes)
regulation of organ growth (10 genes)
negative regulation of organ growth (13 genes)
positive regulation of organ growth (13 genes)
regulation of insulin receptor signaling pathway (13 genes)
negative regulation of insulin receptor signaling pathway (38 genes)
positive regulation of insulin receptor signaling pathway (17 genes)
alpha-beta t cell differentiation (10 genes)
positive regulation of alpha-beta t cell differentiation (10 genes)
positive regulation of alpha-beta t cell proliferation (12 genes)
negative regulation of alpha-beta t cell proliferation (4 genes)
lymphocyte activation (4 genes)
lymphocyte proliferation (5 genes)
tetrahydrofolate metabolic process (8 genes)
tetrahydrofolate biosynthetic process (4 genes)
folic acid metabolic process (5 genes)
female sex differentiation (17 genes)
male sex differentiation (10 genes)
negative regulation of insulin secretion (39 genes)
response to antibiotic (21 genes)
response to arsenic-containing substance (7 genes)
response to cadmium ion (13 genes)
response to copper ion (13 genes)
response to mercury ion (7 genes)
decidualization (18 genes)
muscle cell cellular homeostasis (20 genes)
viral entry into host cell (19 genes)
positive regulation by virus of viral protein levels in host cell (4 genes)
viral budding (6 genes)
protein autophosphorylation (178 genes)
viral mrna export from host cell nucleus (7 genes)
microtubule polymerization (13 genes)
receptor-mediated virion attachment to host cell (4 genes)
regulation of nucleocytoplasmic transport (7 genes)
regulation of protein export from nucleus (8 genes)
negative regulation of protein export from nucleus (6 genes)
positive regulation of protein export from nucleus (18 genes)
positive regulation of rna export from nucleus (5 genes)
lipid phosphorylation (12 genes)
carbohydrate phosphorylation (14 genes)
phospholipid dephosphorylation (12 genes)
filopodium assembly (17 genes)
bone remodeling (18 genes)
regulation of bone remodeling (11 genes)
negative regulation of bone remodeling (4 genes)
phosphatidylinositol phosphorylation (41 genes)
inositol phosphate dephosphorylation (15 genes)
phosphatidylinositol dephosphorylation (21 genes)
positive regulation of saliva secretion (5 genes)
hormone secretion (5 genes)
positive regulation of follicle-stimulating hormone secretion (4 genes)
negative regulation of follicle-stimulating hormone secretion (4 genes)
regulation of hormone secretion (5 genes)
positive regulation of hormone biosynthetic process (5 genes)
positive regulation of hormone secretion (16 genes)
negative regulation of hormone secretion (15 genes)
positive regulation of lipid biosynthetic process (18 genes)
regulation of lipid biosynthetic process (4 genes)
regulation of mitochondrial membrane permeability (11 genes)
secretion (5 genes)
intracellular transport (19 genes)
regulation of neurotransmitter secretion (25 genes)
negative regulation of neurotransmitter secretion (6 genes)
pore complex assembly (6 genes)
ketone body catabolic process (4 genes)
habituation (5 genes)
regulation of response to osmotic stress (5 genes)
vesicle transport along microtubule (16 genes)
mitochondrion transport along microtubule (6 genes)
antigen processing and presentation of peptide antigen (4 genes)
antigen processing and presentation, exogenous lipid antigen via mhc class ib (4 genes)
platelet-derived growth factor receptor signaling pathway (34 genes)
insulin-like growth factor receptor signaling pathway (12 genes)
vascular endothelial growth factor receptor signaling pathway (20 genes)
neurotrophin trk receptor signaling pathway (15 genes)
hepatocyte growth factor receptor signaling pathway (9 genes)
ephrin receptor signaling pathway (31 genes)
tie signaling pathway (4 genes)
phosphatidylinositol-mediated signaling (34 genes)
inositol phosphate-mediated signaling (8 genes)
inositol lipid-mediated signaling (4 genes)
positive regulation of melanin biosynthetic process (6 genes)
regulation of mrna splicing, via spliceosome (10 genes)
negative regulation of mrna splicing, via spliceosome (24 genes)
positive regulation of mrna splicing, via spliceosome (30 genes)
focal adhesion assembly (23 genes)
embryonic eye morphogenesis (6 genes)
developmental pigmentation (16 genes)
eye pigmentation (4 genes)
regulation of developmental pigmentation (7 genes)
chromatin-mediated maintenance of transcription (10 genes)
autophagic cell death (7 genes)
somatic stem cell division (12 genes)
establishment of body hair planar orientation (5 genes)
male germ-line stem cell asymmetric division (4 genes)
astrocyte activation (8 genes)
regulation of fibroblast proliferation (5 genes)
positive regulation of fibroblast proliferation (68 genes)
negative regulation of fibroblast proliferation (36 genes)
behavioral response to cocaine (25 genes)
behavioral response to ethanol (10 genes)
regulation of synaptic plasticity (50 genes)
regulation of neuronal synaptic plasticity (25 genes)
regulation of long-term neuronal synaptic plasticity (32 genes)
positive regulation of long-term neuronal synaptic plasticity (7 genes)
regulation of short-term neuronal synaptic plasticity (18 genes)
golgi vesicle transport (18 genes)
golgi vesicle budding (4 genes)
copi coating of golgi vesicle (4 genes)
copii vesicle coating (7 genes)
plasma membrane to endosome transport (7 genes)
sperm capacitation (29 genes)
eosinophil chemotaxis (18 genes)
macrophage chemotaxis (14 genes)
lymphocyte chemotaxis (28 genes)
elastic fiber assembly (9 genes)
snorna localization (5 genes)
mrna stabilization (23 genes)
positive regulation of receptor-mediated endocytosis (23 genes)
determination of dorsal identity (6 genes)
response to pain (20 genes)
behavioral response to pain (22 genes)
clathrin coat assembly (17 genes)
vesicle docking (20 genes)
vesicle fusion with golgi apparatus (8 genes)
lung alveolus development (50 genes)
positive regulation of isotype switching to ige isotypes (4 genes)
positive regulation of isotype switching to iga isotypes (7 genes)
positive regulation of isotype switching to igg isotypes (15 genes)
immunoglobulin secretion (6 genes)
mitochondrion distribution (8 genes)
intracellular distribution of mitochondria (11 genes)
axial mesoderm development (6 genes)
mesodermal cell differentiation (4 genes)
paraxial mesoderm development (11 genes)
paraxial mesoderm formation (8 genes)
lateral mesoderm development (6 genes)
retinoic acid receptor signaling pathway (16 genes)
regulation of retinoic acid receptor signaling pathway (4 genes)
positive regulation of retinoic acid receptor signaling pathway (5 genes)
negative regulation of retinoic acid receptor signaling pathway (7 genes)
cell development (62 genes)
cell maturation (41 genes)
oogenesis (39 genes)
replication fork protection (8 genes)
enteric nervous system development (14 genes)
sympathetic nervous system development (17 genes)
synaptic vesicle endocytosis (42 genes)
synaptic vesicle transport (19 genes)
anterograde synaptic vesicle transport (17 genes)
maintenance of animal organ identity (5 genes)
regulation of timing of cell differentiation (6 genes)
rhythmic process (131 genes)
circadian behavior (7 genes)
animal organ development (14 genes)
blood vessel morphogenesis (34 genes)
spermatid differentiation (12 genes)
positive regulation of viral process (7 genes)
negative regulation of viral process (5 genes)
lymph node development (31 genes)
spleen development (45 genes)
thymus development (55 genes)
bone marrow development (8 genes)
peyer's patch development (11 genes)
response to steroid hormone (12 genes)
digestive tract morphogenesis (18 genes)
positive regulation of pinocytosis (4 genes)
negative regulation of pinocytosis (5 genes)
embryonic digestive tract morphogenesis (20 genes)
embryonic organ morphogenesis (6 genes)
post-embryonic animal organ morphogenesis (4 genes)
digestive tract development (28 genes)
embryonic digestive tract development (15 genes)
embryonic organ development (38 genes)
notochord morphogenesis (6 genes)
developmental growth (34 genes)
eye morphogenesis (5 genes)
camera-type eye morphogenesis (25 genes)
embryonic camera-type eye morphogenesis (17 genes)
post-embryonic camera-type eye morphogenesis (6 genes)
embryonic morphogenesis (18 genes)
oocyte development (12 genes)
reproductive structure development (12 genes)
multicellular organismal reproductive process (4 genes)
embryonic foregut morphogenesis (12 genes)
embryonic hindgut morphogenesis (4 genes)
skeletal muscle tissue growth (4 genes)
positive regulation of skeletal muscle tissue growth (6 genes)
positive regulation of developmental growth (7 genes)
negative regulation of skeletal muscle tissue development (8 genes)
positive regulation of skeletal muscle tissue development (11 genes)
muscle organ morphogenesis (9 genes)
animal organ formation (7 genes)
anatomical structure formation involved in morphogenesis (22 genes)
smooth muscle cell proliferation (6 genes)
regulation of smooth muscle cell proliferation (11 genes)
positive regulation of smooth muscle cell proliferation (79 genes)
negative regulation of smooth muscle cell proliferation (39 genes)
neuron fate commitment (34 genes)
neuron fate determination (7 genes)
neuron fate specification (17 genes)
neuron development (63 genes)
cell morphogenesis involved in neuron differentiation (13 genes)
collateral sprouting (8 genes)
negative regulation of collateral sprouting (9 genes)
positive regulation of collateral sprouting (10 genes)
axon extension (29 genes)
response to axon injury (18 genes)
regulation of axon regeneration (10 genes)
positive regulation of axon regeneration (8 genes)
negative regulation of axon regeneration (13 genes)
generation of neurons (13 genes)
embryonic cranial skeleton morphogenesis (36 genes)
embryonic neurocranium morphogenesis (7 genes)
embryonic viscerocranium morphogenesis (10 genes)
embryonic skeletal system morphogenesis (58 genes)
skeletal system morphogenesis (52 genes)
embryonic skeletal system development (45 genes)
astrocyte differentiation (10 genes)
oligodendrocyte differentiation (29 genes)
positive regulation of astrocyte differentiation (14 genes)
negative regulation of astrocyte differentiation (17 genes)
regulation of oligodendrocyte differentiation (6 genes)
positive regulation of oligodendrocyte differentiation (25 genes)
negative regulation of oligodendrocyte differentiation (15 genes)
tissue morphogenesis (9 genes)
epidermis morphogenesis (9 genes)
gland development (9 genes)
sebaceous gland development (9 genes)
cardiac muscle tissue development (25 genes)
cardiac muscle fiber development (16 genes)
skeletal muscle fiber development (27 genes)
regulation of skeletal muscle fiber development (5 genes)
positive regulation of skeletal muscle fiber development (8 genes)
smooth muscle tissue development (17 genes)
muscle fiber development (22 genes)
semicircular canal morphogenesis (7 genes)
branching morphogenesis of an epithelial tube (37 genes)
branching morphogenesis of a nerve (5 genes)
mesenchymal cell differentiation (9 genes)
tissue remodeling (14 genes)
calcium ion-regulated exocytosis of neurotransmitter (17 genes)
pronephros development (6 genes)
genitalia development (8 genes)
male genitalia morphogenesis (5 genes)
neuron projection morphogenesis (73 genes)
dendrite morphogenesis (47 genes)
regulation of dendrite morphogenesis (27 genes)
hair follicle maturation (8 genes)
erythrocyte development (25 genes)
inner ear development (61 genes)
positive regulation of axon extension involved in axon guidance (7 genes)
negative regulation of axon extension involved in axon guidance (27 genes)
artery morphogenesis (26 genes)
venous blood vessel morphogenesis (8 genes)
axon extension involved in axon guidance (8 genes)
forebrain morphogenesis (12 genes)
brain morphogenesis (26 genes)
anatomical structure development (24 genes)
cell projection morphogenesis (4 genes)
formation of anatomical boundary (4 genes)
leukemia inhibitory factor signaling pathway (4 genes)
stem cell differentiation (40 genes)
stem cell development (7 genes)
cell motility (21 genes)
homeostasis of number of cells (34 genes)
homeostasis of number of cells within a tissue (40 genes)
chemical homeostasis (4 genes)
peripheral nervous system neuron development (8 genes)
peripheral nervous system neuron axonogenesis (4 genes)
3'-phosphoadenosine 5'-phosphosulfate metabolic process (4 genes)
positive regulation of viral transcription (5 genes)
amyloid-beta metabolic process (16 genes)
arachidonic acid secretion (23 genes)
chondroitin sulfate proteoglycan biosynthetic process (9 genes)
cytokine secretion (8 genes)
hydrogen peroxide biosynthetic process (7 genes)
homocysteine metabolic process (9 genes)
positive regulation of lymphocyte proliferation (10 genes)
negative regulation of lymphocyte proliferation (5 genes)
epithelial cell proliferation (14 genes)
regulation of epithelial cell proliferation (16 genes)
positive regulation of epithelial cell proliferation (77 genes)
negative regulation of epithelial cell proliferation (73 genes)
regulation of mrna processing (13 genes)
negative regulation of defense response to virus (9 genes)
regulation of defense response to virus (8 genes)
negative regulation of defense response to virus by host (4 genes)
regulation of defense response to virus by host (9 genes)
interleukin-1 beta secretion (8 genes)
regulation of cytokine secretion (10 genes)
regulation of protein secretion (14 genes)
negative regulation of protein secretion (15 genes)
negative regulation of cytokine secretion (17 genes)
negative regulation of interleukin-1 secretion (4 genes)
negative regulation of interleukin-1 beta secretion (6 genes)
positive regulation of protein secretion (43 genes)
positive regulation of cytokine secretion (35 genes)
positive regulation of interleukin-1 alpha secretion (4 genes)
positive regulation of interleukin-1 beta secretion (35 genes)
regulation of inflammatory response (64 genes)
negative regulation of inflammatory response (98 genes)
positive regulation of inflammatory response (64 genes)
regulation of peptidyl-tyrosine phosphorylation (22 genes)
positive regulation of peptidyl-tyrosine phosphorylation (110 genes)
negative regulation of peptidyl-tyrosine phosphorylation (17 genes)
chemokine metabolic process (4 genes)
regulation of phagocytosis (11 genes)
negative regulation of phagocytosis (17 genes)
positive regulation of phagocytosis (44 genes)
regulation of neurogenesis (42 genes)
negative regulation of neurogenesis (28 genes)
positive regulation of neurogenesis (39 genes)
regulation of axonogenesis (29 genes)
negative regulation of axonogenesis (14 genes)
positive regulation of axonogenesis (38 genes)
regulation of dendrite development (19 genes)
negative regulation of dendrite morphogenesis (11 genes)
positive regulation of dendrite morphogenesis (32 genes)
regulation of immune response (55 genes)
negative regulation of immune response (12 genes)
positive regulation of immune response (9 genes)
rna destabilization (4 genes)
regulation of catalytic activity (29 genes)
regulation of viral process (6 genes)
regulation of developmental process (5 genes)
regulation of behavior (9 genes)
regulation of insulin secretion (36 genes)
activated t cell proliferation (8 genes)
ion homeostasis (10 genes)
regulation of synapse structure or activity (8 genes)
modulation of chemical synaptic transmission (88 genes)
negative regulation of synaptic transmission (11 genes)
positive regulation of synaptic transmission (34 genes)
regulation of synapse organization (39 genes)
synapse organization (52 genes)
regulation of steroid biosynthetic process (9 genes)
negative regulation of coagulation (6 genes)
positive regulation of coagulation (4 genes)
protein stabilization (162 genes)
defense response to gram-negative bacterium (85 genes)
defense response to gram-positive bacterium (109 genes)
defense response to fungus (18 genes)
progesterone receptor signaling pathway (8 genes)
regulation of calcium-mediated signaling (12 genes)
negative regulation of calcium-mediated signaling (4 genes)
positive regulation of calcium-mediated signaling (23 genes)
t cell receptor signaling pathway (89 genes)
b cell receptor signaling pathway (33 genes)
regulation of t cell receptor signaling pathway (10 genes)
negative regulation of b cell receptor signaling pathway (6 genes)
negative regulation of t cell receptor signaling pathway (18 genes)
positive regulation of b cell receptor signaling pathway (7 genes)
positive regulation of t cell receptor signaling pathway (14 genes)
regulation of t cell activation (10 genes)
negative regulation of t cell activation (18 genes)
negative regulation of b cell activation (8 genes)
positive regulation of t cell activation (19 genes)
positive regulation of b cell activation (9 genes)
white fat cell differentiation (15 genes)
brown fat cell differentiation (35 genes)
nervous system process (55 genes)
regulation of body fluid levels (7 genes)
regulation of blood vessel size (19 genes)
musculoskeletal movement (7 genes)
voluntary musculoskeletal movement (9 genes)
neuromuscular process controlling posture (14 genes)
neuromuscular process controlling balance (60 genes)
cognition (43 genes)
multicellular organismal water homeostasis (7 genes)
intestinal absorption (19 genes)
response to stimulus (217 genes)
leukocyte migration (24 genes)
leukocyte tethering or rolling (19 genes)
diapedesis (4 genes)
neuromuscular process (34 genes)
detection of chemical stimulus involved in sensory perception (5 genes)
detection of light stimulus involved in visual perception (20 genes)
sensory perception of taste (47 genes)
detection of mechanical stimulus involved in sensory perception of sound (18 genes)
sensory perception of bitter taste (6 genes)
sensory perception of sour taste (5 genes)
sensory perception of sweet taste (6 genes)
sensory perception of umami taste (6 genes)
positive chemotaxis (15 genes)
negative chemotaxis (30 genes)
regulation of chemotaxis (7 genes)
positive regulation of chemotaxis (14 genes)
positive regulation of positive chemotaxis (12 genes)
induction of positive chemotaxis (15 genes)
sensory perception of temperature stimulus (4 genes)
sensory perception of light stimulus (12 genes)
thermoception (6 genes)
equilibrioception (7 genes)
detection of temperature stimulus involved in sensory perception of pain (18 genes)
detection of mechanical stimulus involved in sensory perception of pain (20 genes)
detection of mechanical stimulus involved in sensory perception (5 genes)
sensory perception of touch (4 genes)
detection of mechanical stimulus (12 genes)
regulation of lipid catabolic process (4 genes)
negative regulation of lipid catabolic process (16 genes)
positive regulation of lipid catabolic process (7 genes)
regulation of nitric-oxide synthase activity (4 genes)
positive regulation of nitric-oxide synthase activity (17 genes)
negative regulation of nitric-oxide synthase activity (10 genes)
negative regulation of lipoprotein lipase activity (5 genes)
positive regulation of lipoprotein lipase activity (9 genes)
microtubule severing (7 genes)
actin filament severing (7 genes)
barbed-end actin filament capping (16 genes)
actin filament bundle assembly (48 genes)
regulation of immunoglobulin secretion (5 genes)
positive regulation of immunoglobulin secretion (12 genes)
negative regulation of immunoglobulin secretion (5 genes)
chiasma assembly (9 genes)
mrna transport (88 genes)
positive regulation of membrane protein ectodomain proteolysis (15 genes)
negative regulation of membrane protein ectodomain proteolysis (7 genes)
negative regulation of transport (4 genes)
positive regulation of dna metabolic process (6 genes)
negative regulation of lipid biosynthetic process (5 genes)
regulation of small gtpase mediated signal transduction (20 genes)
positive regulation of small gtpase mediated signal transduction (8 genes)
negative regulation of small gtpase mediated signal transduction (6 genes)
nuclear envelope disassembly (5 genes)
'de novo' posttranslational protein folding (4 genes)
chaperone cofactor-dependent protein refolding (30 genes)
regulation of dna binding transcription factor activity (17 genes)
positive regulation of dna binding transcription factor activity (118 genes)
positive regulation of nf-kappab transcription factor activity (124 genes)
positive regulation of helicase activity (6 genes)
regulation of binding (5 genes)
positive regulation of binding (7 genes)
regulation of dna binding (6 genes)
dna ligation involved in dna repair (5 genes)
hepoxilin biosynthetic process (5 genes)
rna polymerase ii transcriptional preinitiation complex assembly (10 genes)
synaptic growth at neuromuscular junction (5 genes)
positive regulation of actin nucleation (11 genes)
chaperone-mediated protein complex assembly (13 genes)
positive regulation of nk t cell activation (6 genes)
positive regulation of nk t cell differentiation (6 genes)
positive regulation of nk t cell proliferation (4 genes)
smooth muscle cell differentiation (18 genes)
striated muscle cell differentiation (14 genes)
regulation of muscle cell differentiation (6 genes)
positive regulation of muscle cell differentiation (7 genes)
regulation of smooth muscle cell differentiation (5 genes)
negative regulation of smooth muscle cell differentiation (7 genes)
positive regulation of smooth muscle cell differentiation (8 genes)
positive regulation of striated muscle cell differentiation (7 genes)
glucose 6-phosphate metabolic process (6 genes)
nuclear export (6 genes)
nuclear import (4 genes)
vitamin transport (7 genes)
protein insertion into mitochondrial membrane (4 genes)
protein insertion into membrane (6 genes)
release of sequestered calcium ion into cytosol (33 genes)
cartilage development (77 genes)
cytoplasmic sequestering of protein (7 genes)
positive regulation of protein transport (6 genes)
regulation of protein transport (12 genes)
negative regulation of protein transport (6 genes)
spindle assembly (31 genes)
regulation of protein metabolic process (13 genes)
positive regulation of protein metabolic process (10 genes)
negative regulation of protein metabolic process (4 genes)
mitotic spindle midzone assembly (7 genes)
protein polymerization (14 genes)
protein oligomerization (79 genes)
protein homooligomerization (235 genes)
protein tetramerization (40 genes)
negative regulation of cellular component movement (6 genes)
positive regulation of cellular component movement (12 genes)
chromosome organization (39 genes)
regulation of release of sequestered calcium ion into cytosol (14 genes)
negative regulation of release of sequestered calcium ion into cytosol (7 genes)
positive regulation of release of sequestered calcium ion into cytosol (34 genes)
protein homotetramerization (86 genes)
protein heterotetramerization (52 genes)
protein heterooligomerization (101 genes)
nuclear pore complex assembly (9 genes)
establishment of spindle orientation (5 genes)
establishment of meiotic spindle localization (4 genes)
centrosome duplication (15 genes)
cell division (384 genes)
regulation of cell division (9 genes)
chromosome separation (5 genes)
mitotic sister chromatid separation (4 genes)
female meiosis chromosome separation (4 genes)
metaphase plate congression (12 genes)
attachment of mitotic spindle microtubules to kinetochore (10 genes)
meiotic cell cycle (158 genes)
negative regulation of cyclic-nucleotide phosphodiesterase activity (4 genes)
negative regulation of hydrolase activity (7 genes)
positive regulation of transferase activity (4 genes)
positive regulation of oxidoreductase activity (9 genes)
negative regulation of oxidoreductase activity (5 genes)
cellular response to potassium ion starvation (4 genes)
kinetochore assembly (12 genes)
kinetochore organization (6 genes)
response to glucocorticoid (30 genes)
negative regulation of neurotrophin trk receptor signaling pathway (5 genes)
positive regulation of neurotrophin trk receptor signaling pathway (9 genes)
neuron apoptotic process (46 genes)
stress-activated mapk cascade (12 genes)
response to corticosterone (10 genes)
interphase microtubule nucleation by interphase microtubule organizing center (6 genes)
regulation of ubiquitin-protein transferase activity (4 genes)
positive regulation of ubiquitin-protein transferase activity (19 genes)
negative regulation of ubiquitin-protein transferase activity (10 genes)
negative regulation of meiotic cell cycle (4 genes)
myoblast proliferation (5 genes)
myoblast migration (10 genes)
regulation of intracellular ph (29 genes)
intracellular ph elevation (5 genes)
maintenance of protein location in nucleus (15 genes)
positive regulation of corticotropin secretion (7 genes)
positive regulation of cortisol secretion (5 genes)
positive regulation of corticotropin-releasing hormone secretion (4 genes)
regulation of cytosolic calcium ion concentration (43 genes)
negative regulation of cytosolic calcium ion concentration (18 genes)
positive regulation of cytosolic calcium ion concentration involved in phospholipase c-activating g-protein coupled signaling pathway (37 genes)
regulation of filopodium assembly (13 genes)
negative regulation of filopodium assembly (4 genes)
positive regulation of filopodium assembly (31 genes)
regulation of stress fiber assembly (16 genes)
regulation of cytoskeleton organization (18 genes)
positive regulation of stress fiber assembly (52 genes)
negative regulation of stress fiber assembly (25 genes)
keratinocyte migration (4 genes)
positive regulation of keratinocyte migration (13 genes)
mitochondrial calcium ion homeostasis (15 genes)
positive regulation of mitochondrial calcium ion concentration (10 genes)
negative regulation of mitochondrial calcium ion concentration (5 genes)
histone h3-k9 methylation (7 genes)
histone h3-k4 methylation (15 genes)
regulation of histone h3-k4 methylation (12 genes)
positive regulation of histone h3-k4 methylation (14 genes)
negative regulation of histone h3-k4 methylation (6 genes)
negative regulation of histone h3-k9 methylation (6 genes)
positive regulation of histone h3-k9 methylation (8 genes)
regulation of neurotransmitter uptake (5 genes)
response to camp (31 genes)
response to calcium ion (50 genes)
exocyst localization (8 genes)
response to electrical stimulus (10 genes)
proteolysis involved in cellular protein catabolic process (60 genes)
protein maturation (21 genes)
defense response to virus (176 genes)
negative regulation of serotonin uptake (4 genes)
actin filament network formation (10 genes)
centrosome localization (19 genes)
golgi localization (6 genes)
mitochondrion localization (8 genes)
nucleus localization (8 genes)
spindle localization (5 genes)
establishment of centrosome localization (5 genes)
maintenance of centrosome location (6 genes)
localization within membrane (5 genes)
membrane disruption in other organism (26 genes)
establishment of golgi localization (8 genes)
actin filament capping (17 genes)
pointed-end actin filament capping (7 genes)
interaction with symbiont (7 genes)
response to other organism (5 genes)
positive regulation of killing of cells of other organism (4 genes)
protein de-adp-ribosylation (4 genes)
regulation of cell cycle (126 genes)
meiotic sister chromatid cohesion, centromeric (5 genes)
actin crosslink formation (10 genes)
positive regulation of nitric-oxide synthase biosynthetic process (19 genes)
negative regulation of nitric-oxide synthase biosynthetic process (7 genes)
response to redox state (8 genes)
positive regulation of cell division (46 genes)
negative regulation of cell division (8 genes)
medium-chain fatty acid metabolic process (4 genes)
medium-chain fatty acid biosynthetic process (6 genes)
medium-chain fatty acid catabolic process (4 genes)
positive regulation of hair follicle development (7 genes)
regulation of synapse structural plasticity (6 genes)
positive regulation of synapse structural plasticity (4 genes)
protein autoubiquitination (67 genes)
killing by host of symbiont cells (5 genes)
regulation of mitochondrial membrane potential (29 genes)
positive regulation of cardioblast differentiation (6 genes)
regulation of focal adhesion assembly (18 genes)
positive regulation of focal adhesion assembly (25 genes)
negative regulation of focal adhesion assembly (16 genes)
regulation of protein kinase b signaling (16 genes)
positive regulation of protein kinase b signaling (102 genes)
negative regulation of protein kinase b signaling (42 genes)
membrane depolarization (21 genes)
regulation of mitochondrial depolarization (5 genes)
positive regulation of mitochondrial depolarization (11 genes)
negative regulation of mitochondrial depolarization (5 genes)
negative regulation of fibrinolysis (7 genes)
positive regulation of fibrinolysis (4 genes)
sulfation (9 genes)
regulation of calcium ion transport (36 genes)
negative regulation of calcium ion transport (12 genes)
positive regulation of calcium ion transport (31 genes)
regulation of sensory perception of pain (45 genes)
synaptic transmission, gabaergic (17 genes)
l-glutamate import (5 genes)
regulation of synapse assembly (13 genes)
negative regulation of synapse assembly (5 genes)
positive regulation of synapse assembly (73 genes)
regulation of synaptic transmission, glutamatergic (27 genes)
negative regulation of synaptic transmission, glutamatergic (11 genes)
positive regulation of synaptic transmission, glutamatergic (34 genes)
regulation of transmission of nerve impulse (4 genes)
negative regulation of transmission of nerve impulse (5 genes)
positive regulation of transmission of nerve impulse (7 genes)
positive regulation of telomerase activity (34 genes)
negative regulation of telomerase activity (14 genes)
regulation of chromosome segregation (10 genes)
positive regulation of chromosome segregation (6 genes)
positive regulation of attachment of spindle microtubules to kinetochore (4 genes)
regulation of attachment of spindle microtubules to kinetochore (7 genes)
regulation of endopeptidase activity (5 genes)
cellular glucuronidation (6 genes)
flavonoid glucuronidation (8 genes)
xenobiotic glucuronidation (8 genes)
muscle cell development (6 genes)
cardiac myofibril assembly (19 genes)
cardiac muscle cell differentiation (35 genes)
cardiac muscle tissue morphogenesis (23 genes)
atrial cardiac muscle tissue morphogenesis (8 genes)
ventricular cardiac muscle tissue morphogenesis (28 genes)
ventricular cardiac muscle cell differentiation (6 genes)
cardiac muscle cell development (16 genes)
ventricular cardiac muscle cell development (9 genes)
phosphate ion homeostasis (10 genes)
chloride ion homeostasis (11 genes)
zinc ion homeostasis (4 genes)
iron ion homeostasis (41 genes)
calcium ion homeostasis (33 genes)
potassium ion homeostasis (17 genes)
sodium ion homeostasis (7 genes)
transmembrane transport (424 genes)
lipid homeostasis (35 genes)
fatty acid homeostasis (15 genes)
phospholipid homeostasis (10 genes)
sterol homeostasis (5 genes)
oxidation-reduction process (686 genes)
regulation of cardiac muscle contraction (16 genes)
relaxation of cardiac muscle (7 genes)
digestive system development (9 genes)
vestibular reflex (5 genes)
sertoli cell differentiation (6 genes)
sertoli cell development (14 genes)
sertoli cell proliferation (7 genes)
synaptic transmission, glycinergic (6 genes)
righting reflex (11 genes)
parathyroid gland development (7 genes)
astrocyte fate commitment (4 genes)
radial glial cell differentiation (6 genes)
bergmann glial cell differentiation (10 genes)
palate development (73 genes)
hard palate development (6 genes)
soft palate development (5 genes)
rhythmic synaptic transmission (4 genes)
convergent extension involved in axis elongation (7 genes)
convergent extension involved in organogenesis (5 genes)
anatomical structure regression (5 genes)
pharyngeal system development (18 genes)
cardiac muscle cell proliferation (17 genes)
pericardium development (10 genes)
retinal bipolar neuron differentiation (9 genes)
retina development in camera-type eye (74 genes)
retina morphogenesis in camera-type eye (14 genes)
regulation of cardiac muscle cell proliferation (9 genes)
negative regulation of cardiac muscle cell proliferation (17 genes)
positive regulation of cardiac muscle cell proliferation (26 genes)
regulation of acrosome reaction (4 genes)
heart contraction (17 genes)
cardiac muscle contraction (51 genes)
positive regulation of protein glycosylation (4 genes)
neurofilament cytoskeleton organization (10 genes)
positive regulation of epithelial cell proliferation involved in wound healing (7 genes)
angiogenesis involved in wound healing (7 genes)
mammary gland involution (6 genes)
positive regulation of apoptotic process involved in mammary gland involution (5 genes)
embryonic retina morphogenesis in camera-type eye (7 genes)
uterus development (20 genes)
vagina development (12 genes)
canonical wnt signaling pathway (79 genes)
wnt signaling pathway, planar cell polarity pathway (15 genes)
micturition (4 genes)
synapse maturation (14 genes)
regulation of resting membrane potential (6 genes)
regulation of postsynaptic membrane potential (20 genes)
excitatory postsynaptic potential (37 genes)
inhibitory postsynaptic potential (14 genes)
membrane hyperpolarization (13 genes)
smooth muscle contraction involved in micturition (4 genes)
relaxation of vascular smooth muscle (8 genes)
auditory receptor cell stereocilium organization (18 genes)
positive regulation of phagocytosis, engulfment (12 genes)
generation of ovulation cycle rhythm (4 genes)
inner ear receptor cell differentiation (9 genes)
auditory receptor cell development (8 genes)
inner ear receptor cell development (10 genes)
inner ear receptor cell stereocilium organization (26 genes)
positive regulation of growth hormone secretion (13 genes)
negative regulation of growth hormone secretion (4 genes)
thyroid-stimulating hormone-secreting cell differentiation (4 genes)
prepulse inhibition (15 genes)
maternal process involved in female pregnancy (11 genes)
embryonic process involved in female pregnancy (9 genes)
maternal process involved in parturition (4 genes)
platelet dense granule organization (4 genes)
urinary bladder development (5 genes)
phospholipase c-activating dopamine receptor signaling pathway (7 genes)
regulation of dopamine receptor signaling pathway (4 genes)
negative regulation of dopamine receptor signaling pathway (4 genes)
positive regulation of dopamine receptor signaling pathway (4 genes)
regulation of timing of neuron differentiation (4 genes)
limb development (40 genes)
limb bud formation (11 genes)
male mating behavior (5 genes)
negative regulation of lipase activity (4 genes)
positive regulation of nuclear-transcribed mrna poly(a) tail shortening (13 genes)
endocardium formation (4 genes)
definitive hemopoiesis (18 genes)
hematopoietic stem cell differentiation (21 genes)
camera-type eye photoreceptor cell differentiation (5 genes)
mesenchymal to epithelial transition (8 genes)
lens induction in camera-type eye (7 genes)
regulation of mitotic spindle organization (17 genes)
negative regulation of cell proliferation involved in contact inhibition (4 genes)
positive regulation of glial cell proliferation (20 genes)
negative regulation of glial cell proliferation (11 genes)
regulation of feeding behavior (16 genes)
positive regulation of transcription initiation from rna polymerase ii promoter (9 genes)
negative regulation of respiratory burst involved in inflammatory response (4 genes)
positive regulation of respiratory burst (4 genes)
cilium assembly (187 genes)
embryonic skeletal joint morphogenesis (12 genes)
positive regulation of ovulation (4 genes)
cilium-dependent cell motility (12 genes)
epithelial cilium movement involved in determination of left/right asymmetry (7 genes)
transdifferentiation (8 genes)
long-term synaptic potentiation (196 genes)
long term synaptic depression (18 genes)
cilium movement involved in cell motility (10 genes)
regulation of cilium beat frequency involved in ciliary motility (6 genes)
regulation of sarcomere organization (4 genes)
positive regulation of sarcomere organization (5 genes)
regulation of phosphatidylinositol dephosphorylation (6 genes)
regulation of membrane repolarization (10 genes)
regulation of ventricular cardiac muscle cell membrane repolarization (18 genes)
regulation of ryanodine-sensitive calcium-release channel activity (8 genes)
negative regulation of ryanodine-sensitive calcium-release channel activity (6 genes)
positive regulation of ryanodine-sensitive calcium-release channel activity (5 genes)
cardiac epithelial to mesenchymal transition (11 genes)
definitive erythrocyte differentiation (4 genes)
head development (18 genes)
head morphogenesis (9 genes)
face development (20 genes)
face morphogenesis (38 genes)
cell chemotaxis (84 genes)
interferon-gamma-mediated signaling pathway (6 genes)
positive regulation of interferon-gamma-mediated signaling pathway (9 genes)
negative regulation of interferon-gamma-mediated signaling pathway (6 genes)
type i interferon signaling pathway (14 genes)
negative regulation of type i interferon-mediated signaling pathway (9 genes)
positive regulation of type i interferon-mediated signaling pathway (11 genes)
regulation of cellular localization (7 genes)
bone trabecula formation (9 genes)
heart trabecula formation (15 genes)
bone development (58 genes)
bone morphogenesis (38 genes)
cartilage development involved in endochondral bone morphogenesis (9 genes)
positive regulation of cell adhesion molecule production (5 genes)
cranial suture morphogenesis (5 genes)
regulation of atrial cardiac muscle cell membrane depolarization (8 genes)
regulation of atrial cardiac muscle cell membrane repolarization (6 genes)
regulation of ventricular cardiac muscle cell membrane depolarization (7 genes)
innervation (23 genes)
axonogenesis involved in innervation (7 genes)
pathway-restricted smad protein phosphorylation (13 genes)
regulation of smad protein import into nucleus (6 genes)
positive regulation of smad protein import into nucleus (13 genes)
negative regulation of smad protein import into nucleus (4 genes)
regulation of pathway-restricted smad protein phosphorylation (4 genes)
negative regulation of pathway-restricted smad protein phosphorylation (13 genes)
smad protein signal transduction (76 genes)
growth hormone receptor signaling pathway (8 genes)
jak-stat cascade involved in growth hormone signaling pathway (4 genes)
calcium ion transport into cytosol (12 genes)
positive regulation of penile erection (9 genes)
cardiac septum morphogenesis (15 genes)
ventricular septum morphogenesis (44 genes)
atrial septum morphogenesis (14 genes)
aorta smooth muscle tissue morphogenesis (5 genes)
muscle tissue morphogenesis (6 genes)
regulation of heart growth (4 genes)
lung morphogenesis (25 genes)
lung vasculature development (9 genes)
lung epithelium development (7 genes)
epithelium development (11 genes)
lung saccule development (8 genes)
lung growth (4 genes)
trachea development (5 genes)
trachea morphogenesis (4 genes)
trachea formation (7 genes)
epithelial tube branching involved in lung morphogenesis (21 genes)
branching involved in prostate gland morphogenesis (7 genes)
mammary gland morphogenesis (4 genes)
branching involved in mammary gland duct morphogenesis (14 genes)
branching involved in salivary gland morphogenesis (15 genes)
bud outgrowth involved in lung branching (4 genes)
bud elongation involved in lung branching (7 genes)
positive regulation of cardiac muscle contraction (9 genes)
negative regulation of gastric acid secretion (7 genes)
lung lobe morphogenesis (8 genes)
prevention of polyspermy (4 genes)
lung-associated mesenchyme development (10 genes)
mesenchyme development (8 genes)
clara cell differentiation (4 genes)
lung epithelial cell differentiation (12 genes)
positive regulation of epithelial cell proliferation involved in lung morphogenesis (7 genes)
type i pneumocyte differentiation (7 genes)
type ii pneumocyte differentiation (7 genes)
prostate gland morphogenesis (5 genes)
prostatic bud formation (4 genes)
prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis (7 genes)
secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development (4 genes)
trachea cartilage development (4 genes)
trachea cartilage morphogenesis (4 genes)
cartilage morphogenesis (7 genes)
skeletal muscle organ development (4 genes)
diaphragm development (8 genes)
respiratory system development (10 genes)
regulation of necroptotic process (6 genes)
negative regulation of necroptotic process (13 genes)
negative regulation of necrotic cell death (6 genes)
negative regulation of cell death (93 genes)
apoptotic process involved in morphogenesis (7 genes)
neuroepithelial cell differentiation (11 genes)
morphogenesis of an epithelial fold (4 genes)
intestinal epithelial cell differentiation (7 genes)
intestinal epithelial cell development (13 genes)
multicellular organismal iron ion homeostasis (11 genes)
chondroblast differentiation (5 genes)
mammary placode formation (4 genes)
mammary gland duct morphogenesis (8 genes)
adipose tissue development (33 genes)
fat pad development (7 genes)
regulation of vesicle-mediated transport (5 genes)
regulation of er to golgi vesicle-mediated transport (6 genes)
mammary gland epithelial cell differentiation (13 genes)
salivary gland cavitation (6 genes)
epithelial cell proliferation involved in salivary gland morphogenesis (7 genes)
regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling (5 genes)
dichotomous subdivision of terminal units involved in salivary gland branching (5 genes)
embryonic placenta morphogenesis (7 genes)
branching involved in labyrinthine layer morphogenesis (10 genes)
placenta blood vessel development (13 genes)
ureteric bud formation (5 genes)
epithelial-mesenchymal cell signaling (4 genes)
negative regulation of prostatic bud formation (4 genes)
regulation of branching involved in prostate gland morphogenesis (7 genes)
regulation of morphogenesis of a branching structure (4 genes)
regulation of branching involved in salivary gland morphogenesis (4 genes)
regulation of ribonuclease activity (12 genes)
cell differentiation involved in embryonic placenta development (6 genes)
trophoblast giant cell differentiation (19 genes)
spongiotrophoblast differentiation (6 genes)
chorio-allantoic fusion (10 genes)
labyrinthine layer development (12 genes)
spongiotrophoblast layer development (7 genes)
labyrinthine layer morphogenesis (4 genes)
labyrinthine layer blood vessel development (26 genes)
chorionic trophoblast cell differentiation (6 genes)
intestinal epithelial structure maintenance (4 genes)
positive regulation of inositol phosphate biosynthetic process (8 genes)
prostate gland growth (10 genes)
prostate gland epithelium morphogenesis (12 genes)
epithelial cell differentiation involved in prostate gland development (6 genes)
epithelial cell maturation involved in prostate gland development (4 genes)
mammary gland branching involved in thelarche (6 genes)
mammary gland alveolus development (19 genes)
epithelial cell proliferation involved in mammary gland duct elongation (5 genes)
positive regulation of mast cell chemotaxis (7 genes)
positive regulation of response to cytokine stimulus (7 genes)
negative regulation of response to cytokine stimulus (5 genes)
regulation of branching involved in mammary gland duct morphogenesis (6 genes)
mammary duct terminal end bud growth (7 genes)
regulation of androgen receptor signaling pathway (11 genes)
negative regulation of androgen receptor signaling pathway (16 genes)
negative regulation of epithelial cell proliferation involved in prostate gland development (7 genes)
hair follicle placode formation (5 genes)
inactivation of x chromosome by genetic imprinting (4 genes)
regulation of canonical wnt signaling pathway (30 genes)
ciliary receptor clustering involved in smoothened signaling pathway (4 genes)
smoothened signaling pathway involved in dorsal/ventral neural tube patterning (7 genes)
lymphatic endothelial cell differentiation (6 genes)
artery development (9 genes)
venous blood vessel development (5 genes)
arterial endothelial cell differentiation (4 genes)
regulation of transcription involved in cell fate commitment (6 genes)
positive regulation of meiosis i (5 genes)
positive regulation of macrophage cytokine production (8 genes)
cardiac cell fate determination (4 genes)
cardiac muscle cell fate commitment (5 genes)
cardiac vascular smooth muscle cell differentiation (9 genes)
cardiac vascular smooth muscle cell development (5 genes)
endocardial cell differentiation (5 genes)
regulation of gene silencing by mirna (8 genes)
negative regulation of gene silencing by mirna (5 genes)
regulation of gene silencing (11 genes)
embryonic heart tube left/right pattern formation (5 genes)
left/right pattern formation (9 genes)
coronary vasculature development (41 genes)
coronary vasculature morphogenesis (5 genes)
angiogenesis involved in coronary vascular morphogenesis (6 genes)
vasculogenesis involved in coronary vascular morphogenesis (15 genes)
coronary artery morphogenesis (10 genes)
kidney morphogenesis (15 genes)
dendritic spine development (20 genes)
dendritic spine morphogenesis (21 genes)
regulation of dendritic spine development (7 genes)
positive regulation of dendritic spine development (38 genes)
negative regulation of dendritic spine development (7 genes)
regulation of dendritic spine morphogenesis (17 genes)
negative regulation of dendritic spine morphogenesis (8 genes)
positive regulation of dendritic spine morphogenesis (24 genes)
common bile duct development (7 genes)
gall bladder development (4 genes)
positive regulation of mrna catabolic process (15 genes)
membrane organization (21 genes)
membrane fusion (21 genes)
establishment of endothelial barrier (17 genes)
eyelid development in camera-type eye (15 genes)
epithelial cell differentiation involved in mammary gland alveolus development (4 genes)
endodermal digestive tract morphogenesis (4 genes)
visceral serous pericardium development (10 genes)
positive regulation of cartilage development (13 genes)
negative regulation of cartilage development (10 genes)
uterus morphogenesis (6 genes)
regulation of wound healing (4 genes)
negative regulation of wound healing (13 genes)
cell growth involved in cardiac muscle cell development (12 genes)
positive regulation of cell growth involved in cardiac muscle cell development (15 genes)
negative regulation of cell growth involved in cardiac muscle cell development (11 genes)
somite development (16 genes)
sclerotome development (4 genes)
iris morphogenesis (7 genes)
ciliary body morphogenesis (5 genes)
chaperone-mediated protein folding (23 genes)
negative regulation of histone h3-k27 methylation (4 genes)
positive regulation of histone h3-k27 methylation (5 genes)
regulation of sequestering of zinc ion (6 genes)
regulation of protein tyrosine kinase activity (4 genes)
positive regulation of protein tyrosine kinase activity (31 genes)
negative regulation of protein tyrosine kinase activity (11 genes)
regulation of proteasomal protein catabolic process (16 genes)
endothelial tube morphogenesis (6 genes)
pulmonary artery morphogenesis (6 genes)
mrna destabilization (12 genes)
3'-utr-mediated mrna destabilization (18 genes)
regulation of insulin secretion involved in cellular response to glucose stimulus (24 genes)
negative regulation of insulin secretion involved in cellular response to glucose stimulus (18 genes)
mammary gland epithelium development (7 genes)
fungiform papilla formation (4 genes)
paramesonephric duct development (5 genes)
retina vasculature development in camera-type eye (17 genes)
retina vasculature morphogenesis in camera-type eye (8 genes)
cornea development in camera-type eye (8 genes)
retinal blood vessel morphogenesis (6 genes)
cardiac neural crest cell development involved in outflow tract morphogenesis (6 genes)
bmp signaling pathway involved in heart development (6 genes)
notch signaling involved in heart development (7 genes)
renal tubule morphogenesis (5 genes)
cardiac conduction (11 genes)
neural precursor cell proliferation (16 genes)
testosterone biosynthetic process (5 genes)
determination of heart left/right asymmetry (11 genes)
heart trabecula morphogenesis (9 genes)
positive regulation of transcription from rna polymerase ii promoter in response to heat stress (6 genes)
positive regulation of transcription from rna polymerase ii promoter in response to hypoxia (6 genes)
negative regulation of transcription from rna polymerase ii promoter in response to hypoxia (4 genes)
establishment of skin barrier (23 genes)
connective tissue development (5 genes)
reproductive system development (6 genes)
protein localization to lysosome (8 genes)
regulation of type b pancreatic cell proliferation (8 genes)
t follicular helper cell differentiation (4 genes)
hematopoietic stem cell homeostasis (8 genes)
protein localization to cilium (30 genes)
sympathetic ganglion development (9 genes)
trigeminal ganglion development (5 genes)
axon development (14 genes)
calcium activated phospholipid scrambling (5 genes)
calcium activated phosphatidylserine scrambling (4 genes)
calcium activated phosphatidylcholine scrambling (5 genes)
calcium activated galactosylceramide scrambling (5 genes)
pri-mirna transcription by rna polymerase ii (10 genes)
glycolytic process through fructose-6-phosphate (5 genes)
canonical glycolysis (5 genes)
pharyngeal arch artery morphogenesis (10 genes)
regulation of protein complex stability (8 genes)
mitochondrial ribosome assembly (5 genes)
spontaneous neurotransmitter secretion (5 genes)
mitochondrial acetyl-coa biosynthetic process from pyruvate (8 genes)
parkin-mediated stimulation of mitophagy in response to mitochondrial depolarization (5 genes)
protein lipidation involved in autophagosome assembly (4 genes)
motor learning (5 genes)
motor behavior (5 genes)
camkk-ampk signaling cascade (4 genes)
dense core granule priming (4 genes)
telomeric dna-containing double minutes formation (4 genes)
telomeric d-loop disassembly (8 genes)
antimicrobial humoral immune response mediated by antimicrobial peptide (71 genes)
intracellular protein transmembrane transport (4 genes)
protein-dna complex assembly (6 genes)
neuron cellular homeostasis (14 genes)
intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress (37 genes)
vacuolar proton-transporting v-type atpase complex assembly (4 genes)
negative regulation of glucagon secretion (6 genes)
positive regulation of glucagon secretion (4 genes)
chemokine-mediated signaling pathway (48 genes)
negative regulation of chemokine-mediated signaling pathway (5 genes)
interleukin-6-mediated signaling pathway (10 genes)
ciliary neurotrophic factor-mediated signaling pathway (5 genes)
mitochondrial translational elongation (4 genes)
regulation of mitochondrial translation (7 genes)
positive regulation of mitochondrial translation (18 genes)
response to uv-a (4 genes)
negative regulation of adiponectin secretion (4 genes)
enamel mineralization (9 genes)
negative regulation of biomineral tissue development (5 genes)
positive regulation of biomineral tissue development (5 genes)
positive regulation of enamel mineralization (4 genes)
kynurenine metabolic process (4 genes)
chromosome organization involved in meiotic cell cycle (7 genes)
meiotic attachment of telomere to nuclear envelope (6 genes)
protein localization to chromosome, telomeric region (9 genes)
establishment of protein localization to telomere (5 genes)
regulation of establishment of protein localization (10 genes)
protein trimerization (7 genes)
protein homotrimerization (38 genes)
protein heterotrimerization (19 genes)
protein poly-adp-ribosylation (6 genes)
protein auto-adp-ribosylation (4 genes)
t cell apoptotic process (4 genes)
negative regulation of t cell apoptotic process (12 genes)
positive regulation of t cell apoptotic process (6 genes)
negative regulation of activation-induced cell death of t cells (5 genes)
thymocyte apoptotic process (5 genes)
regulation of thymocyte apoptotic process (4 genes)
negative regulation of thymocyte apoptotic process (10 genes)
positive regulation of thymocyte apoptotic process (6 genes)
actin-mediated cell contraction (6 genes)
positive regulation of mucus secretion (10 genes)
peptidyl-serine dephosphorylation (10 genes)
necroptotic process (15 genes)
pyroptosis (23 genes)
axonemal dynein complex assembly (10 genes)
n-acylethanolamine metabolic process (4 genes)
renal sodium ion absorption (4 genes)
renal water absorption (4 genes)
sarcoplasmic reticulum calcium ion transport (6 genes)
cellular response to hydrogen peroxide (48 genes)
lens fiber cell differentiation (11 genes)
lens fiber cell development (9 genes)
lens fiber cell morphogenesis (6 genes)
g1 to g0 transition (4 genes)
negative regulation of g0 to g1 transition (5 genes)
positive regulation of g0 to g1 transition (4 genes)
thyroid hormone transport (5 genes)
triglyceride homeostasis (28 genes)
negative regulation of fat cell proliferation (5 genes)
actin polymerization-dependent cell motility (4 genes)
hepatocyte differentiation (11 genes)
erk1 and erk2 cascade (29 genes)
regulation of erk1 and erk2 cascade (32 genes)
negative regulation of erk1 and erk2 cascade (79 genes)
positive regulation of erk1 and erk2 cascade (248 genes)
harderian gland development (4 genes)
cellular response to cold (8 genes)
nucleotide-binding oligomerization domain containing 2 signaling pathway (7 genes)
regulation of oligodendrocyte progenitor proliferation (5 genes)
positive regulation of oligodendrocyte progenitor proliferation (6 genes)
positive regulation of uterine smooth muscle contraction (9 genes)
rrna base methylation (6 genes)
response to oxygen levels (46 genes)
thrombin-activated receptor signaling pathway (8 genes)
interleukin-1-mediated signaling pathway (15 genes)
regulation of microtubule cytoskeleton organization (35 genes)
cholesterol import (6 genes)
calcium ion import (27 genes)
platelet aggregation (25 genes)
protein kinase c signaling (17 genes)
protein k63-linked ubiquitination (38 genes)
protein k63-linked deubiquitination (25 genes)
histone h2a k63-linked deubiquitination (4 genes)
response to fatty acid (8 genes)
histone h3-k36 demethylation (9 genes)
response to interleukin-1 (22 genes)
cadmium ion transmembrane transport (5 genes)
mitochondrion morphogenesis (24 genes)
protein localization to mitochondrion (9 genes)
calcium ion transmembrane transport (98 genes)
dendrite self-avoidance (15 genes)
regulation of protein processing (11 genes)
transepithelial transport (6 genes)
positive regulation of mast cell proliferation (4 genes)
proteasome regulatory particle assembly (4 genes)
response to cholesterol (6 genes)
histone h3-k27 methylation (6 genes)
d-aspartate import (5 genes)
phosphorylation of rna polymerase ii c-terminal domain (7 genes)
heterochromatin organization (4 genes)
bicellular tight junction assembly (26 genes)
caveola assembly (4 genes)
dehydroascorbic acid transport (4 genes)
aggresome assembly (7 genes)
response to growth factor (6 genes)
negative regulation of bile acid biosynthetic process (4 genes)
positive regulation of bile acid biosynthetic process (4 genes)
positive regulation of protein exit from endoplasmic reticulum (10 genes)
regulation of glycogen metabolic process (4 genes)
regulation of calcineurin-nfat signaling cascade (7 genes)
negative regulation of calcineurin-nfat signaling cascade (15 genes)
positive regulation of calcineurin-nfat signaling cascade (18 genes)
rna polymerase iii transcriptional preinitiation complex assembly (4 genes)
uv-damage excision repair (12 genes)
histone h3 deacetylation (14 genes)
histone h4 deacetylation (12 genes)
crd-mediated mrna stabilization (6 genes)
3'-utr-mediated mrna stabilization (14 genes)
protein k48-linked ubiquitination (56 genes)
dephosphorylation of rna polymerase ii c-terminal domain (5 genes)
neutrophil mediated killing of gram-negative bacterium (7 genes)
nuclear-transcribed mrna catabolic process, no-go decay (4 genes)
protein localization to endoplasmic reticulum (4 genes)
protein localization to endoplasmic reticulum exit site (5 genes)
protein k11-linked ubiquitination (30 genes)
left/right axis specification (12 genes)
nadph oxidation (7 genes)
neuron death (14 genes)
nuclear mrna surveillance (9 genes)
cut catabolic process (4 genes)
nuclear polyadenylation-dependent rrna catabolic process (6 genes)
nuclear polyadenylation-dependent trna catabolic process (5 genes)
nuclear polyadenylation-dependent mrna catabolic process (4 genes)
histone mrna catabolic process (11 genes)
polyadenylation-dependent snorna 3'-end processing (6 genes)
protein k48-linked deubiquitination (29 genes)
regulation of cell cycle arrest (4 genes)
negative regulation of cell cycle arrest (20 genes)
positive regulation of cell cycle arrest (28 genes)
protein localization to chromatin (14 genes)
establishment of protein localization to chromatin (8 genes)
protein localization to juxtaparanode region of axon (5 genes)
cellular response to biotic stimulus (4 genes)
cellular response to misfolded protein (6 genes)
cellular response to molecule of bacterial origin (6 genes)
cellular response to lipopolysaccharide (234 genes)
cellular response to lipoteichoic acid (11 genes)
cellular response to peptidoglycan (7 genes)
cellular response to muramyl dipeptide (9 genes)
cellular response to acid chemical (7 genes)
cellular response to amino acid stimulus (141 genes)
cellular response to leucine (10 genes)
cellular response to phenylalanine (25 genes)
cellular response to antibiotic (5 genes)
cellular response to inorganic substance (48 genes)
cellular response to arsenic-containing substance (8 genes)
cellular response to electrical stimulus (10 genes)
cellular response to mechanical stimulus (65 genes)
cellular response to cadmium ion (23 genes)
cellular response to calcium ion (77 genes)
cellular response to copper ion (13 genes)
cellular response to iron ion (7 genes)
cellular response to lithium ion (13 genes)
cellular response to magnesium ion (5 genes)
cellular response to manganese ion (4 genes)
cellular response to zinc ion (12 genes)
cellular response to retinoic acid (49 genes)
cellular response to organic substance (43 genes)
cellular response to alkaloid (4 genes)
cellular response to caffeine (6 genes)
cellular response to nicotine (5 genes)
cellular response to atp (12 genes)
cellular response to camp (39 genes)
cellular response to cgmp (7 genes)
cellular response to carbohydrate stimulus (4 genes)
cellular response to fructose stimulus (4 genes)
cellular response to glucose stimulus (50 genes)
skeletal muscle acetylcholine-gated channel clustering (8 genes)
cellular response to cytokine stimulus (34 genes)
cellular response to interferon-gamma (99 genes)
cellular response to interleukin-1 (55 genes)
cellular response to interleukin-4 (24 genes)
cellular response to interleukin-6 (15 genes)
cellular response to tumor necrosis factor (104 genes)
cellular response to dsrna (6 genes)
cellular response to exogenous dsrna (16 genes)
cellular response to ethanol (27 genes)
cellular response to growth factor stimulus (53 genes)
cellular response to epidermal growth factor stimulus (40 genes)
cellular response to gonadotropin stimulus (6 genes)
cellular response to follicle-stimulating hormone stimulus (9 genes)
cellular response to parathyroid hormone stimulus (4 genes)
cellular response to peptide hormone stimulus (12 genes)
cellular response to corticotropin-releasing hormone stimulus (4 genes)
cellular response to prostaglandin e stimulus (14 genes)
cellular response to steroid hormone stimulus (11 genes)
cellular response to glucocorticoid stimulus (17 genes)
cellular response to estrogen stimulus (37 genes)
cellular response to estradiol stimulus (24 genes)
cellular response to testosterone stimulus (8 genes)
cellular response to cholesterol (9 genes)
cellular response to fatty acid (10 genes)
cellular response to low-density lipoprotein particle stimulus (13 genes)
cellular response to organic cyclic compound (53 genes)
cellular response to histamine (8 genes)
hematopoietic stem cell proliferation (16 genes)
positive regulation of histone h3-k14 acetylation (5 genes)
cellular response to hydroperoxide (7 genes)
cellular response to hyperoxia (5 genes)
cellular response to hypoxia (68 genes)
protein localization to chromosome, centromeric region (6 genes)
cellular response to xenobiotic stimulus (4 genes)
cellular response to ph (9 genes)
cellular response to acidic ph (9 genes)
cellular response to osmotic stress (10 genes)
cellular hyperosmotic response (4 genes)
cellular hyperosmotic salinity response (18 genes)
cellular hypotonic response (7 genes)
cellular response to radiation (4 genes)
cellular response to ionizing radiation (32 genes)
cellular response to gamma radiation (28 genes)
cellular response to x-ray (8 genes)
cellular response to light stimulus (12 genes)
cellular response to uv-b (8 genes)
cellular response to uv-c (6 genes)
cellular response to fluid shear stress (12 genes)
cellular response to laminar fluid shear stress (6 genes)
cellular response to temperature stimulus (4 genes)
cellular response to heparin (4 genes)
genetic imprinting (9 genes)
semaphorin-plexin signaling pathway (37 genes)
protein localization to centrosome (20 genes)
dopaminergic neuron differentiation (27 genes)
diphosphoinositol polyphosphate metabolic process (5 genes)
response to dexamethasone (4 genes)
cellular response to dexamethasone stimulus (29 genes)
histone h3-k27 demethylation (5 genes)
response to transforming growth factor beta (9 genes)
cellular response to transforming growth factor beta stimulus (50 genes)
protein ufmylation (5 genes)
zinc ii ion transmembrane transport (14 genes)
zinc ii ion transmembrane import (7 genes)
otic vesicle development (6 genes)
vocalization behavior (13 genes)
positive regulation of transforming growth factor beta production (7 genes)
negative regulation of monocyte chemotactic protein-1 production (8 genes)
positive regulation of monocyte chemotactic protein-1 production (11 genes)
negative regulation of chemokine (c-c motif) ligand 5 production (4 genes)
positive regulation of chemokine (c-c motif) ligand 5 production (5 genes)
negative regulation of smooth muscle cell chemotaxis (4 genes)
positive regulation of mononuclear cell migration (4 genes)
commissural neuron axon guidance (13 genes)
cellular response to indole-3-methanol (5 genes)
organic substance metabolic process (6 genes)
basement membrane organization (12 genes)
er-associated misfolded protein catabolic process (12 genes)
cellular response to diacyl bacterial lipopeptide (4 genes)
response to nitric oxide (4 genes)
cellular response to nitric oxide (11 genes)
transcriptional activation by promoter-enhancer looping (4 genes)
nuclear membrane organization (4 genes)
cellular response to bmp stimulus (33 genes)
endoplasmic reticulum tubular network organization (9 genes)
endoplasmic reticulum tubular network formation (4 genes)
podosome assembly (4 genes)
positive regulation of podosome assembly (14 genes)
potassium ion transmembrane transport (103 genes)
tail-anchored membrane protein insertion into er membrane (6 genes)
tnfsf11-mediated signaling pathway (5 genes)
mitotic cell cycle arrest (14 genes)
positive regulation of cell proliferation in bone marrow (10 genes)
negative regulation of apoptotic process in bone marrow (5 genes)
cellular response to catecholamine stimulus (6 genes)
cellular response to epinephrine stimulus (9 genes)
adrenergic receptor signaling pathway (5 genes)
adenylate cyclase-activating adrenergic receptor signaling pathway (21 genes)
odontoblast differentiation (4 genes)
protein localization to adherens junction (4 genes)
dna biosynthetic process (18 genes)
regulation of protein serine/threonine kinase activity (6 genes)
negative regulation of protein serine/threonine kinase activity (10 genes)
positive regulation of protein serine/threonine kinase activity (35 genes)
determination of digestive tract left/right asymmetry (6 genes)
determination of liver left/right asymmetry (6 genes)
regulation of cohesin loading (4 genes)
protein deubiquitination involved in ubiquitin-dependent protein catabolic process (4 genes)
multivesicular body sorting pathway (8 genes)
renal system development (23 genes)
nephron development (8 genes)
glomerulus vasculature development (4 genes)
proximal tubule development (4 genes)
glomerular visceral epithelial cell development (7 genes)
comma-shaped body morphogenesis (4 genes)
s-shaped body morphogenesis (6 genes)
metanephric mesenchyme development (5 genes)
stem cell proliferation (6 genes)
regulation of stem cell proliferation (12 genes)
glomerulus morphogenesis (6 genes)
positive regulation of ureteric bud formation (4 genes)
positive regulation of mesenchymal to epithelial transition involved in metanephros morphogenesis (4 genes)
glomerular visceral epithelial cell differentiation (6 genes)
positive regulation of glomerular mesangial cell proliferation (6 genes)
condensed mesenchymal cell proliferation (4 genes)
epithelial cell fate commitment (4 genes)
metanephric mesenchymal cell differentiation (4 genes)
mesonephric tubule development (5 genes)
nephric duct morphogenesis (5 genes)
ureter development (6 genes)
ureter smooth muscle cell differentiation (4 genes)
negative regulation of mesenchymal cell proliferation (6 genes)
metanephric collecting duct development (6 genes)
metanephric nephron development (4 genes)
metanephric distal convoluted tubule development (4 genes)
metanephric nephron tubule formation (4 genes)
regulation of metanephric nephron tubule epithelial cell differentiation (6 genes)
chaperone-mediated protein transport (9 genes)
intrinsic apoptotic signaling pathway by p53 class mediator (26 genes)
rescue of stalled ribosome (5 genes)
cardiovascular system development (19 genes)
blood coagulation, fibrin clot formation (7 genes)
plus-end-directed vesicle transport along microtubule (4 genes)
organelle transport along microtubule (7 genes)
signal transduction involved in g2 dna damage checkpoint (11 genes)
response to intra-s dna damage checkpoint signaling (4 genes)
ammonium transmembrane transport (4 genes)
embryonic skeletal joint development (4 genes)
positive regulation of secondary heart field cardioblast proliferation (5 genes)
seminiferous tubule development (20 genes)
t-helper 17 cell lineage commitment (8 genes)
terminal button organization (4 genes)
hepatocyte proliferation (6 genes)
endothelial cell apoptotic process (4 genes)
clathrin-dependent endocytosis (18 genes)
caveolin-mediated endocytosis (5 genes)
reactive oxygen species metabolic process (38 genes)
establishment of protein localization to organelle (5 genes)
interleukin-4 secretion (6 genes)
interleukin-8 secretion (4 genes)
interleukin-17 secretion (4 genes)
interferon-gamma secretion (5 genes)
establishment of protein localization to mitochondrion (7 genes)
maintenance of protein location in mitochondrion (5 genes)
protein localization to membrane (24 genes)
protein localization to plasma membrane (139 genes)
neutrophil extravasation (8 genes)
lamellipodium morphogenesis (7 genes)
multinuclear osteoclast differentiation (4 genes)
osteoclast fusion (6 genes)
lymphocyte migration (4 genes)
t cell migration (7 genes)
cellular response to hydroxyurea (9 genes)
cellular response to cisplatin (4 genes)
cellular response to camptothecin (5 genes)
autophagy of host cells involved in interaction with symbiont (8 genes)
ires-dependent viral translational initiation (10 genes)
viral translational termination-reinitiation (5 genes)
mrna methylation (11 genes)
dna demethylation (12 genes)
histone h3-k4 trimethylation (16 genes)
protein k6-linked ubiquitination (8 genes)
extracellular matrix assembly (6 genes)
cardiac muscle cell action potential (4 genes)
cardiac muscle cell action potential involved in contraction (13 genes)
regulation of cardiac muscle cell contraction (17 genes)
ventricular cardiac muscle cell action potential (16 genes)
membrane repolarization (9 genes)
membrane depolarization during action potential (16 genes)
membrane repolarization during action potential (8 genes)
membrane depolarization during cardiac muscle cell action potential (6 genes)
atrial cardiac muscle cell action potential (7 genes)
sa node cell action potential (6 genes)
av node cell action potential (4 genes)
regulation of cardiac muscle cell membrane potential (9 genes)
membrane depolarization during av node cell action potential (5 genes)
membrane depolarization during sa node cell action potential (4 genes)
bundle of his cell-purkinje myocyte adhesion involved in cell communication (6 genes)
regulation of heart rate by cardiac conduction (33 genes)
primitive streak formation (11 genes)
regulation of neutrophil chemotaxis (4 genes)
positive regulation of neutrophil chemotaxis (24 genes)
positive regulation of monocyte chemotaxis (19 genes)
negative regulation of monocyte chemotaxis (6 genes)
positive regulation of steroid hormone biosynthetic process (6 genes)
regulation of protein kinase c signaling (6 genes)
positive regulation of protein kinase c signaling (9 genes)
negative regulation of protein kinase c signaling (4 genes)
regulation of tubulin deacetylation (4 genes)
positive regulation of cell migration involved in sprouting angiogenesis (21 genes)
negative regulation of cell migration involved in sprouting angiogenesis (17 genes)
positive regulation of protein homodimerization activity (12 genes)
negative regulation of protein homodimerization activity (4 genes)
positive regulation of mapkkk cascade by fibroblast growth factor receptor signaling pathway (6 genes)
negative regulation of inclusion body assembly (7 genes)
negative regulation of canonical wnt signaling pathway (119 genes)
positive regulation of extracellular matrix disassembly (10 genes)
cochlea development (23 genes)
cochlea morphogenesis (28 genes)
cargo loading into copii-coated vesicle (13 genes)
copii-coated vesicle budding (7 genes)
regulation of synapse maturation (5 genes)
positive regulation of synapse maturation (10 genes)
actin filament branching (5 genes)
epithelial cell-cell adhesion (10 genes)
regulation of mitochondrial fission (8 genes)
positive regulation of mitochondrial fission (15 genes)
establishment of protein localization to membrane (9 genes)
cellular sphingolipid homeostasis (4 genes)
golgi to lysosome transport (8 genes)
golgi ribbon formation (13 genes)
establishment of epithelial cell polarity (15 genes)
golgi disassembly (6 genes)
golgi reassembly (4 genes)
regulation of golgi inheritance (4 genes)
regulation of establishment of planar polarity (5 genes)
planar cell polarity pathway involved in neural tube closure (13 genes)
regulation of cholesterol metabolic process (14 genes)
positive regulation of kidney development (5 genes)
negative regulation of pancreatic juice secretion (5 genes)
positive regulation of branching involved in ureteric bud morphogenesis (23 genes)
negative regulation of branching involved in ureteric bud morphogenesis (4 genes)
positive regulation of chemokine secretion (11 genes)
positive regulation of release of cytochrome c from mitochondria (27 genes)
negative regulation of release of cytochrome c from mitochondria (20 genes)
protein localization to nuclear pore (4 genes)
regulation of triglyceride metabolic process (9 genes)
negative regulation of triglyceride metabolic process (5 genes)
regulation of metaphase plate congression (5 genes)
positive regulation of arachidonic acid secretion (6 genes)
positive regulation of histone h4 acetylation (4 genes)
wnt signaling pathway involved in somitogenesis (4 genes)
negative regulation of mitochondrial fission (6 genes)
positive regulation of inclusion body assembly (5 genes)
positive regulation of canonical wnt signaling pathway (92 genes)
regulation of mitotic cell cycle spindle assembly checkpoint (5 genes)
positive regulation of mitotic cell cycle spindle assembly checkpoint (7 genes)
positive regulation of fibroblast growth factor production (4 genes)
negative regulation of fibroblast growth factor production (5 genes)
positive regulation of somatostatin secretion (4 genes)
positive regulation of peptide hormone secretion (7 genes)
regulation of calcium ion import (8 genes)
positive regulation of calcium ion import (20 genes)
negative regulation of calcium ion import (9 genes)
cytoskeletal anchoring at nuclear membrane (9 genes)
positive regulation of wound healing (15 genes)
nucleic acid phosphodiester bond hydrolysis (9 genes)
spindle assembly involved in meiosis (4 genes)
mitotic spindle assembly (35 genes)
regulation of protein deacetylation (4 genes)
positive regulation of protein deacetylation (6 genes)
positive regulation of protein targeting to membrane (28 genes)
negative regulation of protein targeting to membrane (6 genes)
positive regulation of intracellular protein transport (24 genes)
negative regulation of oxidative phosphorylation (9 genes)
regulation of platelet aggregation (5 genes)
negative regulation of platelet aggregation (13 genes)
regulation of brown fat cell differentiation (5 genes)
positive regulation of brown fat cell differentiation (11 genes)
negative regulation of cell aging (7 genes)
negative regulation of cholesterol efflux (5 genes)
phagosome maturation (6 genes)
phagosome acidification (5 genes)
phagosome-lysosome fusion (7 genes)
negative regulation of excitatory postsynaptic potential (10 genes)
cellular senescence (19 genes)
replicative senescence (9 genes)
oxidative stress-induced premature senescence (4 genes)
protein localization to nuclear envelope (4 genes)
pyrimidine nucleotide-sugar transmembrane transport (6 genes)
rna phosphodiester bond hydrolysis (4 genes)
rna phosphodiester bond hydrolysis, endonucleolytic (10 genes)
rna phosphodiester bond hydrolysis, exonucleolytic (7 genes)
glomerular visceral epithelial cell migration (6 genes)
actin filament reorganization (8 genes)
establishment of endothelial intestinal barrier (12 genes)
regulation of membrane permeability (7 genes)
ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway (5 genes)
activation of gtpase activity (77 genes)
modulation of age-related behavioral decline (7 genes)
cellular response to oxygen-glucose deprivation (4 genes)
t-circle formation (5 genes)
scarna localization to cajal body (5 genes)
telomerase rna stabilization (4 genes)
acetylcholine receptor signaling pathway (9 genes)
regulation of plasma lipoprotein particle levels (6 genes)
energy homeostasis (38 genes)
cellular response to granulocyte macrophage colony-stimulating factor stimulus (14 genes)
dendritic cell differentiation (11 genes)
mature conventional dendritic cell differentiation (4 genes)
type b pancreatic cell apoptotic process (4 genes)
dendritic spine organization (4 genes)
dendritic spine maintenance (9 genes)
response to thyroid hormone (4 genes)
cellular response to thyroid hormone stimulus (8 genes)
ductus arteriosus closure (6 genes)
vascular smooth muscle cell development (6 genes)
synaptic vesicle clustering (8 genes)
craniofacial suture morphogenesis (8 genes)
postsynaptic membrane assembly (9 genes)
presynaptic membrane assembly (9 genes)
postsynaptic density assembly (5 genes)
gamma-aminobutyric acid receptor clustering (4 genes)
ampa glutamate receptor clustering (5 genes)
nmda glutamate receptor clustering (6 genes)
gephyrin clustering involved in postsynaptic density assembly (4 genes)
neuroligin clustering involved in postsynaptic membrane assembly (5 genes)
postsynaptic density protein 95 clustering (7 genes)
receptor localization to synapse (13 genes)
neuronal stem cell population maintenance (23 genes)
positive regulation of inhibitory postsynaptic potential (9 genes)
circadian regulation of translation (4 genes)
ruffle assembly (13 genes)
amelogenesis (10 genes)
apoptotic signaling pathway (47 genes)
extrinsic apoptotic signaling pathway (44 genes)
extrinsic apoptotic signaling pathway in absence of ligand (38 genes)
intrinsic apoptotic signaling pathway (23 genes)
execution phase of apoptosis (14 genes)
activation of cysteine-type endopeptidase activity (13 genes)
cellular response to gonadotropin-releasing hormone (4 genes)
cellular response to toxic substance (6 genes)
amyloid-beta clearance (11 genes)
mitochondrial respiratory chain supercomplex assembly (4 genes)
self proteolysis (10 genes)
hepatocyte apoptotic process (14 genes)
iron ion import (5 genes)
programmed necrotic cell death (9 genes)
plasma membrane tubulation (14 genes)
mitochondrial outer membrane permeabilization (7 genes)
neutrophil clearance (4 genes)
autophagosome maturation (20 genes)
establishment of sertoli cell barrier (5 genes)
interneuron axon guidance (8 genes)
hypoxia-inducible factor-1alpha signaling pathway (4 genes)
liver regeneration (11 genes)
protein maturation by iron-sulfur cluster transfer (9 genes)
supramolecular fiber organization (11 genes)
dendrite extension (11 genes)
neuron projection guidance (8 genes)
sympathetic neuron projection extension (4 genes)
sympathetic neuron projection guidance (4 genes)
regulation of vesicle size (4 genes)
receptor localization to non-motile cilium (7 genes)
necroptotic signaling pathway (6 genes)
calcium ion transmembrane import into cytosol (7 genes)
potassium ion export across plasma membrane (15 genes)
l-arginine import across plasma membrane (5 genes)
amylin receptor signaling pathway (6 genes)
double-strand break repair via classical nonhomologous end joining (4 genes)
calcineurin-mediated signaling (6 genes)
regulation of blood vessel diameter (11 genes)
positive regulation of blood vessel diameter (34 genes)
negative regulation of blood vessel diameter (12 genes)
histone h3-k27 trimethylation (8 genes)
de novo centriole assembly involved in multi-ciliated epithelial cell differentiation (6 genes)
cellular response to virus (24 genes)
cell-cell adhesion (134 genes)
cation transmembrane transport (11 genes)
anion transmembrane transport (14 genes)
neurotransmitter loading into synaptic vesicle (6 genes)
calcium ion import across plasma membrane (9 genes)
l-glutamate import across plasma membrane (6 genes)
sodium ion import across plasma membrane (14 genes)
positive regulation of the force of heart contraction (5 genes)
cell-cell adhesion via plasma-membrane adhesion molecules (10 genes)
positive regulation of mitophagy in response to mitochondrial depolarization (5 genes)
response to mitochondrial depolarisation (7 genes)
pre-mrna cleavage required for polyadenylation (12 genes)
modulation of excitatory postsynaptic potential (6 genes)
postsynaptic neurotransmitter receptor internalization (9 genes)
modification of postsynaptic actin cytoskeleton (10 genes)
neurotransmitter receptor transport, endosome to postsynaptic membrane (10 genes)
regulation of av node cell action potential (4 genes)
regulation of neuronal action potential (5 genes)
regulation of cardiac muscle cell action potential involved in regulation of contraction (5 genes)
regulation of ventricular cardiac muscle cell action potential (11 genes)
membrane repolarization during atrial cardiac muscle cell action potential (4 genes)
membrane repolarization during ventricular cardiac muscle cell action potential (4 genes)
retrograde trans-synaptic signaling by trans-synaptic protein complex (5 genes)
neurotransmitter receptor transport, postsynaptic endosome to lysosome (9 genes)
regulation of postsynaptic neurotransmitter receptor activity (14 genes)
dendritic transport of messenger ribonucleoprotein complex (6 genes)
exocytic insertion of neurotransmitter receptor to postsynaptic membrane (8 genes)
postsynaptic neurotransmitter receptor diffusion trapping (12 genes)
postsynaptic actin cytoskeleton organization (15 genes)
nmda selective glutamate receptor signaling pathway (4 genes)
regulation of postsynaptic neurotransmitter receptor internalization (23 genes)
regulation of presynaptic cytosolic calcium ion concentration (9 genes)
presynaptic dense core vesicle exocytosis (6 genes)
postsynapse to nucleus signaling pathway (15 genes)
trans-synaptic signaling by endocannabinoid, modulating synaptic transmission (4 genes)
synaptic membrane adhesion (25 genes)
modification of synaptic structure (4 genes)
regulation of postsynaptic cytosolic calcium ion concentration (5 genes)
regulation of translation at postsynapse, modulating synaptic transmission (4 genes)
neurotransmitter receptor internalization (10 genes)
regulation of neurotransmitter receptor activity (4 genes)
anterograde axonal protein transport (8 genes)
neurotransmitter receptor localization to postsynaptic specialization membrane (16 genes)
positive regulation of adenylate cyclase-activating g-protein coupled receptor signaling pathway (4 genes)
negative regulation of adenylate cyclase-activating g-protein coupled receptor signaling pathway (7 genes)
intermembrane lipid transfer (7 genes)
cellular response to oxidised low-density lipoprotein particle stimulus (8 genes)
neuron projection arborization (4 genes)
negative regulation of serine-type endopeptidase activity (5 genes)
positive regulation of dendrite development (18 genes)
regulation of cytokine production involved in inflammatory response (5 genes)
negative regulation of cytokine production involved in inflammatory response (15 genes)
positive regulation of cytokine production involved in inflammatory response (19 genes)
positive regulation of protein kinase c activity (7 genes)
regulation of substrate adhesion-dependent cell spreading (5 genes)
negative regulation of substrate adhesion-dependent cell spreading (13 genes)
positive regulation of substrate adhesion-dependent cell spreading (36 genes)
regulation of ruffle assembly (8 genes)
negative regulation of ruffle assembly (6 genes)
positive regulation of ruffle assembly (15 genes)
positive regulation of interleukin-2 secretion (7 genes)
regulation of cellular response to insulin stimulus (4 genes)
positive regulation of cellular response to insulin stimulus (5 genes)
positive regulation of peptidyl-tyrosine autophosphorylation (7 genes)
positive regulation of g1/s transition of mitotic cell cycle (31 genes)
positive regulation of endoplasmic reticulum unfolded protein response (5 genes)
negative regulation of histone h3-k9 trimethylation (7 genes)
positive regulation of histone h3-k9 trimethylation (4 genes)
negative regulation of execution phase of apoptosis (7 genes)
positive regulation of execution phase of apoptosis (10 genes)
negative regulation of receptor binding (7 genes)
positive regulation of receptor binding (8 genes)
positive regulation of hyaluronan biosynthetic process (5 genes)
positive regulation of nuclear-transcribed mrna catabolic process, deadenylation-dependent decay (16 genes)
negative regulation of bone mineralization involved in bone maturation (4 genes)
nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry (4 genes)
negative regulation of interleukin-6 secretion (9 genes)
regulation of protein localization to nucleus (7 genes)
negative regulation of protein localization to nucleus (14 genes)
positive regulation of protein localization to nucleus (27 genes)
negative regulation of clathrin-dependent endocytosis (4 genes)
negative regulation of oocyte maturation (4 genes)
negative regulation of mesenchymal cell apoptotic process involved in metanephros development (4 genes)
positive regulation of amyloid-beta clearance (5 genes)
negative regulation of nlrp3 inflammasome complex assembly (5 genes)
positive regulation of nlrp3 inflammasome complex assembly (7 genes)
regulation of synaptic vesicle endocytosis (15 genes)
positive regulation of synaptic vesicle endocytosis (12 genes)
positive regulation of rig-i signaling pathway (8 genes)
positive regulation of dna-directed dna polymerase activity (8 genes)
regulation of long-term synaptic potentiation (14 genes)
negative regulation of long-term synaptic potentiation (10 genes)
positive regulation of long-term synaptic potentiation (25 genes)
negative regulation of mrna polyadenylation (8 genes)
positive regulation of mrna polyadenylation (5 genes)
regulation of cellular response to oxidative stress (7 genes)
negative regulation of defense response to bacterium (4 genes)
positive regulation of defense response to bacterium (6 genes)
regulation of long term synaptic depression (5 genes)
positive regulation of long term synaptic depression (8 genes)
negative regulation of g1/s transition of mitotic cell cycle by negative regulation of transcription from rna polymerase ii promoter (5 genes)
positive regulation of phospholipase c-activating g-protein coupled receptor signaling pathway (4 genes)
regulation of p38mapk cascade (7 genes)
positive regulation of p38mapk cascade (27 genes)
regulation of vascular endothelial growth factor signaling pathway (4 genes)
negative regulation of vascular endothelial growth factor signaling pathway (6 genes)
positive regulation of vascular endothelial growth factor signaling pathway (6 genes)
regulation of membrane depolarization during cardiac muscle cell action potential (4 genes)
regulation of potassium ion transmembrane transporter activity (6 genes)
negative regulation of potassium ion transmembrane transporter activity (7 genes)
positive regulation of potassium ion transmembrane transporter activity (7 genes)
negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway (7 genes)
positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway (7 genes)
negative regulation of autophagosome maturation (4 genes)
positive regulation of autophagosome maturation (8 genes)
insulin metabolic process (7 genes)
negative regulation of trophoblast cell migration (5 genes)
positive regulation of trophoblast cell migration (5 genes)
neural crest cell migration involved in autonomic nervous system development (7 genes)
positive regulation of extracellular matrix assembly (4 genes)
regulation of neuron death (19 genes)
negative regulation of neuron death (65 genes)
positive regulation of neuron death (40 genes)
regulation of nik/nf-kappab signaling (11 genes)
negative regulation of nik/nf-kappab signaling (21 genes)
positive regulation of nik/nf-kappab signaling (53 genes)
negative regulation of hydrogen peroxide-mediated programmed cell death (4 genes)
positive regulation of hydrogen peroxide-mediated programmed cell death (4 genes)
regulation of potassium ion transmembrane transport (16 genes)
negative regulation of potassium ion transmembrane transport (11 genes)
positive regulation of potassium ion transmembrane transport (16 genes)
regulation of voltage-gated calcium channel activity (9 genes)
negative regulation of voltage-gated calcium channel activity (10 genes)
positive regulation of voltage-gated calcium channel activity (11 genes)
negative regulation of lymphangiogenesis (4 genes)
positive regulation of transcription from rna polymerase ii promoter involved in cellular response to chemical stimulus (8 genes)
negative regulation of mitophagy (5 genes)
regulation of hematopoietic progenitor cell differentiation (4 genes)
positive regulation of dna demethylation (4 genes)
negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning (6 genes)
nucleoside transmembrane transport (8 genes)
calcium ion export (4 genes)
regulation of mitotic spindle assembly (21 genes)
positive regulation of histone deacetylase activity (6 genes)
positive regulation of myoblast fusion (22 genes)
regulation of signal transduction by p53 class mediator (11 genes)
negative regulation of signal transduction by p53 class mediator (6 genes)
positive regulation of signal transduction by p53 class mediator (6 genes)
negative regulation of proteasomal protein catabolic process (12 genes)
positive regulation of proteasomal protein catabolic process (12 genes)
negative regulation of transcription of nucleolar large rrna by rna polymerase i (4 genes)
positive regulation of transcription of nucleolar large rrna by rna polymerase i (10 genes)
regulation of high voltage-gated calcium channel activity (5 genes)
negative regulation of high voltage-gated calcium channel activity (5 genes)
positive regulation of high voltage-gated calcium channel activity (5 genes)
positive regulation of cellular respiration (4 genes)
regulation of cell junction assembly (4 genes)
positive regulation of calcium-transporting atpase activity (5 genes)
regulation of relaxation of cardiac muscle (4 genes)
diadenosine pentaphosphate catabolic process (5 genes)
diadenosine hexaphosphate catabolic process (5 genes)
adenosine 5'-(hexahydrogen pentaphosphate) catabolic process (5 genes)
positive regulation of protein acetylation (10 genes)
regulation of mitotic cell cycle phase transition (9 genes)
negative regulation of mitotic cell cycle phase transition (4 genes)
toxin transport (37 genes)
positive regulation of amyloid-beta formation (11 genes)
regulation of cilium assembly (20 genes)
negative regulation of cilium assembly (7 genes)
positive regulation of hematopoietic stem cell proliferation (6 genes)
regulation of hematopoietic stem cell differentiation (5 genes)
regulation of extrinsic apoptotic signaling pathway via death domain receptors (5 genes)
negative regulation of extrinsic apoptotic signaling pathway via death domain receptors (27 genes)
positive regulation of extrinsic apoptotic signaling pathway via death domain receptors (15 genes)
negative regulation of peptidyl-cysteine s-nitrosylation (6 genes)
positive regulation of flagellated sperm motility (7 genes)
regulation of mitochondrial membrane permeability involved in apoptotic process (8 genes)
negative regulation of mitochondrial membrane permeability involved in apoptotic process (4 genes)
positive regulation of mitochondrial membrane permeability involved in apoptotic process (4 genes)
regulation of organelle assembly (4 genes)
negative regulation of intrinsic apoptotic signaling pathway in response to dna damage by p53 class mediator (11 genes)
regulation of oxidative stress-induced intrinsic apoptotic signaling pathway (4 genes)
negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway (10 genes)
positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway (4 genes)
negative regulation of viral release from host cell (15 genes)
positive regulation of viral release from host cell (10 genes)
negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress (4 genes)
negative regulation of intrinsic apoptotic signaling pathway in response to dna damage (15 genes)
positive regulation of intrinsic apoptotic signaling pathway in response to dna damage (10 genes)
negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway (17 genes)
positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway (8 genes)
negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator (8 genes)
positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator (5 genes)
regulation of delayed rectifier potassium channel activity (6 genes)
negative regulation of delayed rectifier potassium channel activity (11 genes)
positive regulation of delayed rectifier potassium channel activity (4 genes)
apoptotic process involved in blood vessel morphogenesis (6 genes)
regulation of chromatin organization (5 genes)
semaphorin-plexin signaling pathway involved in neuron projection guidance (4 genes)
semaphorin-plexin signaling pathway involved in axon guidance (12 genes)
regulation of sodium ion transmembrane transport (7 genes)
protein localization to cell junction (5 genes)
positive regulation of mrna binding (6 genes)
negative regulation of amyloid-beta formation (12 genes)
positive regulation of stem cell population maintenance (9 genes)
positive regulation of protein localization to synapse (5 genes)
chloride transmembrane transport (41 genes)
regulation of apoptotic dna fragmentation (4 genes)
positive regulation of apoptotic dna fragmentation (4 genes)
regulation of organelle transport along microtubule (5 genes)
regulation of calcium ion transmembrane transport via high voltage-gated calcium channel (5 genes)
regulation of intracellular signal transduction (5 genes)
positive regulation of intracellular signal transduction (8 genes)
protein localization to nucleolus (7 genes)
hydrogen ion transmembrane transport (31 genes)
regulation of axon guidance (8 genes)
negative regulation of interferon-gamma secretion (5 genes)
positive regulation of interferon-gamma secretion (12 genes)
positive regulation of chondrocyte proliferation (7 genes)
positive regulation of interferon-alpha secretion (11 genes)
apoptotic process involved in development (4 genes)
positive regulation of cell cycle g2/m phase transition (6 genes)
late endosome to lysosome transport (4 genes)
negative regulation of cell cycle g1/s phase transition (6 genes)
positive regulation of cell cycle g1/s phase transition (9 genes)
negative regulation of protein localization to microtubule (5 genes)
positive regulation of non-motile cilium assembly (13 genes)
negative regulation of pri-mirna transcription by rna polymerase ii (15 genes)
positive regulation of pri-mirna transcription by rna polymerase ii (33 genes)
regulation of postsynaptic density protein 95 clustering (4 genes)
negative regulation of autophagosome assembly (12 genes)
positive regulation of protein polyubiquitination (8 genes)
positive regulation of tau-protein kinase activity (5 genes)
regulation of dendritic spine maintenance (4 genes)
negative regulation of dendritic spine maintenance (4 genes)
positive regulation of dendritic spine maintenance (5 genes)
positive regulation of er to golgi vesicle-mediated transport (4 genes)
positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process (6 genes)
positive regulation of protein localization to early endosome (8 genes)
mitotic telomere maintenance via semi-conservative replication (4 genes)
positive regulation of protein deubiquitination (4 genes)
regulation of rna polymerase ii regulatory region sequence-specific dna binding (6 genes)
negative regulation of rna polymerase ii regulatory region sequence-specific dna binding (5 genes)
negative regulation of leukocyte cell-cell adhesion (6 genes)
positive regulation of leukocyte cell-cell adhesion (4 genes)
positive regulation of proteolysis involved in cellular protein catabolic process (4 genes)
regulation of extracellular matrix organization (6 genes)
positive regulation of protein lipidation (4 genes)
positive regulation of er-associated ubiquitin-dependent protein catabolic process (5 genes)
regulation of protein localization to plasma membrane (24 genes)
negative regulation of protein localization to plasma membrane (18 genes)
positive regulation of protein localization to plasma membrane (55 genes)
regulation of establishment of endothelial barrier (6 genes)
positive regulation of establishment of endothelial barrier (5 genes)
regulation of autophagy of mitochondrion (8 genes)
regulation of calcium ion transmembrane transport (7 genes)
negative regulation of oxidative stress-induced cell death (5 genes)
negative regulation of oxidative stress-induced neuron death (17 genes)
negative regulation of hydrogen peroxide-induced cell death (14 genes)
negative regulation of hydrogen peroxide-induced neuron death (6 genes)
regulation of protein targeting to mitochondrion (6 genes)
negative regulation of protein targeting to mitochondrion (5 genes)
melanosome assembly (5 genes)
positive regulation of leukocyte tethering or rolling (5 genes)
negative regulation of cardiac muscle hypertrophy in response to stress (6 genes)
positive regulation of tumor necrosis factor-mediated signaling pathway (7 genes)
positive regulation of sodium ion export across plasma membrane (4 genes)
positive regulation of potassium ion import (7 genes)
negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway (5 genes)
positive regulation of regulated secretory pathway (5 genes)
negative regulation of bicellular tight junction assembly (4 genes)
cellular response to dopamine (7 genes)
l-ornithine transmembrane transport (5 genes)
regulation of golgi organization (11 genes)
protein localization to basolateral plasma membrane (4 genes)
negative regulation of cellular protein catabolic process (7 genes)
positive regulation of cellular protein catabolic process (14 genes)
positive regulation of endoplasmic reticulum tubular network organization (4 genes)
negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway (4 genes)
positive regulation of adherens junction organization (6 genes)
positive regulation of synaptic vesicle recycling (4 genes)
negative regulation of reactive oxygen species biosynthetic process (14 genes)
positive regulation of reactive oxygen species biosynthetic process (14 genes)
regulation of torc1 signaling (6 genes)
protein localization to ciliary membrane (4 genes)
positive regulation of lactation (28 genes)
positive regulation of membrane tubulation (4 genes)
positive regulation of exosomal secretion (16 genes)
protein localization to photoreceptor outer segment (8 genes)
positive regulation of extracellular exosome assembly (4 genes)
positive regulation of protein localization to cilium (4 genes)
negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis (4 genes)
positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis (9 genes)
positive regulation of histamine secretion by mast cell (5 genes)
positive regulation of autophagy of mitochondrion (5 genes)
protein localization to cytoplasmic stress granule (7 genes)
negative regulation of sprouting angiogenesis (10 genes)
positive regulation of sprouting angiogenesis (19 genes)
negative regulation of wound healing, spreading of epidermal cells (4 genes)
regulation of aerobic respiration (7 genes)
positive regulation of i-kappab phosphorylation (5 genes)
positive regulation of centriole elongation (4 genes)
negative regulation of p38mapk cascade (7 genes)
regulation of cardiac conduction (6 genes)
negative regulation of production of mirnas involved in gene silencing by mirna (9 genes)
positive regulation of production of mirnas involved in gene silencing by mirna (9 genes)
negative regulation of voltage-gated potassium channel activity (5 genes)
positive regulation of voltage-gated potassium channel activity (10 genes)
regulation of cellular protein localization (17 genes)
positive regulation of cellular protein localization (7 genes)
positive regulation of aorta morphogenesis (4 genes)
negative regulation of dendrite extension (4 genes)
positive regulation of dendrite extension (27 genes)
positive regulation of oxidative phosphorylation (5 genes)
positive regulation of perk-mediated unfolded protein response (4 genes)
negative regulation of endoplasmic reticulum stress-induced eif2 alpha phosphorylation (5 genes)
cellular response to sodium arsenite (4 genes)
positive regulation of protein targeting to mitochondrion (6 genes)
monounsaturated fatty acid biosynthetic process (4 genes)
positive regulation of microglial cell activation (7 genes)
negative regulation of eating behavior (5 genes)
positive regulation of eating behavior (5 genes)
negative regulation of cyclin-dependent protein kinase activity (4 genes)
positive regulation of epithelial cell apoptotic process (4 genes)
positive regulation of sensory perception of pain (9 genes)
negative regulation of retrograde protein transport, er to cytosol (8 genes)
positive regulation of adipose tissue development (8 genes)
positive regulation of basement membrane assembly involved in embryonic body morphogenesis (5 genes)
negative regulation of torc1 signaling (16 genes)
positive regulation of torc1 signaling (10 genes)
positive regulation of erad pathway (5 genes)
cellular response to forskolin (64 genes)
negative regulation of telomere capping (5 genes)
positive regulation of telomere capping (18 genes)
negative regulation of telomere maintenance via telomere lengthening (7 genes)
positive regulation of telomere maintenance via telomere lengthening (5 genes)
mannose trimming involved in glycoprotein erad pathway (4 genes)
cellular response to angiotensin (7 genes)
positive regulation of skeletal muscle acetylcholine-gated channel clustering (4 genes)
positive regulation of xenophagy (8 genes)
negative regulation of tubulin deacetylation (6 genes)
folic acid import across plasma membrane (5 genes)
negative regulation of tumor necrosis factor secretion (9 genes)
positive regulation of tumor necrosis factor secretion (17 genes)
protein localization to ciliary transition zone (6 genes)
positive regulation of lipophagy (5 genes)
positive regulation of microtubule binding (4 genes)
positive regulation of protein import (4 genes)
cellular response to ionomycin (5 genes)
response to amyloid-beta (5 genes)
cellular response to amyloid-beta (25 genes)
glucose transmembrane transport (15 genes)
negative regulation of ubiquitin protein ligase activity (10 genes)
positive regulation of ubiquitin protein ligase activity (7 genes)
negative regulation of vascular smooth muscle cell proliferation (18 genes)
positive regulation of vascular smooth muscle cell proliferation (34 genes)
regulation of ampa glutamate receptor clustering (4 genes)
positive regulation of protein localization to nucleolus (6 genes)
negative regulation of vascular associated smooth muscle cell migration (6 genes)
positive regulation of vascular associated smooth muscle cell migration (18 genes)
positive regulation of protein localization to cell cortex (5 genes)
positive regulation of protein localization to centrosome (7 genes)
positive regulation of nmda glutamate receptor activity (4 genes)
nlrp1 inflammasome complex assembly (4 genes)
positive regulation of core promoter binding (6 genes)
positive regulation of establishment of protein localization to telomere (10 genes)
regulation of telomerase rna localization to cajal body (4 genes)
positive regulation of telomerase rna localization to cajal body (4 genes)
cranial skeletal system development (7 genes)
positive regulation of stat cascade (5 genes)
positive regulation of autophagy of mitochondrion in response to mitochondrial depolarization (5 genes)
planar cell polarity pathway involved in axon guidance (7 genes)
midbrain dopaminergic neuron differentiation (9 genes)
positive regulation of establishment of protein localization (5 genes)
positive regulation of epithelial to mesenchymal transition involved in endocardial cushion formation (5 genes)
autophagosome organization (4 genes)
positive regulation of metallopeptidase activity (5 genes)
negative regulation of vascular smooth muscle cell differentiation (5 genes)
negative regulation of interleukin-17 secretion (4 genes)
negative regulation of guanyl-nucleotide exchange factor activity (6 genes)
positive regulation of double-strand break repair via homologous recombination (7 genes)
positive regulation of connective tissue replacement (4 genes)
regulation of modification of synaptic structure (5 genes)
regulation of modification of postsynaptic actin cytoskeleton (8 genes)
telomerase holoenzyme complex assembly (4 genes)
positive regulation of protein localization to kinetochore (4 genes)
positive regulation of fc-gamma receptor signaling pathway involved in phagocytosis (4 genes)
positive regulation of vascular associated smooth muscle cell apoptotic process (7 genes)
non-motile cilium assembly (52 genes)
positive regulation of fertilization (5 genes)
negative regulation of vascular endothelial cell proliferation (7 genes)
positive regulation of vascular endothelial cell proliferation (15 genes)
regulation of presynapse assembly (31 genes)
positive regulation of dna methylation (4 genes)
positive regulation of calcium ion import across plasma membrane (4 genes)
negative regulation of protection from non-homologous end joining at telomere (4 genes)
positive regulation of spindle assembly (5 genes)
amyloid fibril formation (9 genes)
inhibition of cysteine-type endopeptidase activity involved in apoptotic process (7 genes)
calcium ion export across plasma membrane (5 genes)
anterograde neuronal dense core vesicle transport (4 genes)
lens fiber cell apoptotic process (4 genes)
cellular response to nerve growth factor stimulus (26 genes)
retrograde transport, endosome to plasma membrane (12 genes)
neuron projection extension (26 genes)
protein k33-linked deubiquitination (4 genes)
exosomal secretion (5 genes)
cellular response to leucine starvation (10 genes)
peptidyl-tyrosine dephosphorylation involved in inactivation of protein kinase activity (5 genes)
neutrophil migration (7 genes)
cellular response to insulin-like growth factor stimulus (8 genes)
embryonic brain development (19 genes)
adrenomedullin receptor signaling pathway (4 genes)
replication-born double-strand break repair via sister chromatid exchange (6 genes)
cellular response to brain-derived neurotrophic factor stimulus (4 genes)
positive regulation of transcription from rna polymerase ii promoter in response to endoplasmic reticulum stress (11 genes)
negative regulation of transcription from rna polymerase ii promoter in response to endoplasmic reticulum stress (5 genes)
mrna pseudouridine synthesis (5 genes)
pirna biosynthetic process (5 genes)
neuron projection maintenance (10 genes)
potassium ion import across plasma membrane (40 genes)
protein k69-linked ufmylation (5 genes)
protein sialylation (5 genes)
endoplasmic reticulum tubular network membrane organization (4 genes)
cellular response to leukemia inhibitory factor (312 genes)
adaptive thermogenesis (7 genes)
cellular response to chemokine (13 genes)
response to amino acid starvation (4 genes)
atp generation from poly-adp-d-ribose (4 genes)
regulation of dna damage checkpoint (10 genes)
negative regulation of dna damage checkpoint (4 genes)
regulation of protein localization to cell surface (9 genes)
negative regulation of protein localization to cell surface (12 genes)
positive regulation of protein localization to cell surface (20 genes)
positive regulation of male gonad development (7 genes)
regulation of stem cell division (11 genes)
regulation of stem cell population maintenance (17 genes)
negative regulation of double-strand break repair via homologous recombination (15 genes)
regulation of g1/s transition of mitotic cell cycle (18 genes)
negative regulation of cell-cell adhesion mediated by cadherin (5 genes)
positive regulation of cell-cell adhesion mediated by cadherin (8 genes)
negative regulation of non-canonical wnt signaling pathway (6 genes)
positive regulation of non-canonical wnt signaling pathway (6 genes)
regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process (4 genes)
negative regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process (19 genes)
positive regulation of protein ubiquitination involved in ubiquitin-dependent protein catabolic process (14 genes)
negative regulation of cortisol biosynthetic process (4 genes)
positive regulation of wnt signaling pathway, planar cell polarity pathway (7 genes)
negative regulation of macrophage apoptotic process (6 genes)
regulation of establishment of cell polarity (16 genes)
negative regulation of cysteine-type endopeptidase activity (4 genes)
regulation of removal of superoxide radicals (5 genes)
negative regulation of g1/s transition of mitotic cell cycle (43 genes)
positive regulation of cell proliferation involved in heart morphogenesis (4 genes)
positive regulation of dna-templated transcription, initiation (8 genes)
regulation of cell motility (10 genes)
negative regulation of cell motility (13 genes)
positive regulation of cell motility (16 genes)
negative regulation of dendrite development (10 genes)
negative regulation of branching morphogenesis of a nerve (4 genes)
regulation of neural precursor cell proliferation (13 genes)
negative regulation of neural precursor cell proliferation (14 genes)
positive regulation of neural precursor cell proliferation (28 genes)
positive regulation of anoikis (6 genes)
positive regulation of rrna processing (9 genes)
regulation of actin cytoskeleton reorganization (15 genes)
positive regulation of actin cytoskeleton reorganization (19 genes)
negative regulation of feeding behavior (11 genes)
positive regulation of feeding behavior (9 genes)
negative regulation of male germ cell proliferation (4 genes)
regulation of fibroblast apoptotic process (4 genes)
negative regulation of fibroblast apoptotic process (9 genes)
positive regulation of fibroblast apoptotic process (7 genes)
negative regulation of receptor activity (6 genes)
positive regulation of receptor activity (7 genes)
negative regulation of dna biosynthetic process (15 genes)
positive regulation of myoblast proliferation (8 genes)
regulation of myoblast proliferation (4 genes)
regulation of synaptic vesicle exocytosis (73 genes)
negative regulation of synaptic vesicle exocytosis (5 genes)
positive regulation of ceramide biosynthetic process (4 genes)
regulation of nmda receptor activity (17 genes)
regulation of ampa receptor activity (25 genes)
positive regulation of t-helper 17 type immune response (5 genes)
negative regulation of t-helper 17 cell differentiation (9 genes)
positive regulation of t-helper 17 cell differentiation (5 genes)
negative regulation of glucocorticoid receptor signaling pathway (6 genes)
positive regulation of glucocorticoid receptor signaling pathway (5 genes)
regulation of terminal button organization (4 genes)
positive regulation of chemokine (c-x-c motif) ligand 2 production (7 genes)
positive regulation of acrosome reaction (10 genes)
regulation of hepatocyte proliferation (4 genes)
negative regulation of hepatocyte proliferation (5 genes)
positive regulation of hepatocyte proliferation (7 genes)
negative regulation of endothelial cell apoptotic process (31 genes)
positive regulation of endothelial cell apoptotic process (20 genes)
regulation of clathrin-dependent endocytosis (8 genes)
positive regulation of clathrin-dependent endocytosis (5 genes)
positive regulation of dna topoisomerase (atp-hydrolyzing) activity (4 genes)
regulation of reactive oxygen species metabolic process (27 genes)
negative regulation of reactive oxygen species metabolic process (28 genes)
positive regulation of reactive oxygen species metabolic process (31 genes)
negative regulation of mesoderm development (5 genes)
positive regulation of neutrophil extravasation (8 genes)
regulation of lamellipodium morphogenesis (4 genes)
positive regulation of lamellipodium morphogenesis (7 genes)
negative regulation of lymphocyte migration (4 genes)
positive regulation of t cell migration (19 genes)
positive regulation of apoptotic cell clearance (4 genes)
negative regulation of protein neddylation (4 genes)
positive regulation of excitatory postsynaptic potential (34 genes)
positive regulation of glycogen (starch) synthase activity (5 genes)
regulation of camp-dependent protein kinase activity (4 genes)
negative regulation of camp-dependent protein kinase activity (8 genes)
positive regulation of camp-dependent protein kinase activity (5 genes)
negative regulation of interleukin-8 secretion (6 genes)
positive regulation of interleukin-8 secretion (13 genes)
positive regulation of hepatic stellate cell activation (6 genes)
positive regulation of natural killer cell chemotaxis (4 genes)
positive regulation of blood vessel remodeling (4 genes)
positive regulation of dendritic cell chemotaxis (7 genes)
positive regulation of t cell costimulation (4 genes)
regulation of entry of bacterium into host cell (4 genes)
positive regulation of gastrulation (7 genes)
positive regulation of endothelial cell chemotaxis to fibroblast growth factor (4 genes)
positive regulation of t-helper 2 cell cytokine production (6 genes)
positive regulation of t-helper 1 cell cytokine production (6 genes)
negative regulation of cd4-positive, alpha-beta t cell proliferation (8 genes)
positive regulation of cd8-positive, alpha-beta t cell proliferation (4 genes)
positive regulation of dna biosynthetic process (21 genes)
positive regulation of atp-dependent microtubule motor activity, plus-end-directed (4 genes)
negative regulation of platelet-derived growth factor receptor-beta signaling pathway (4 genes)
positive regulation of arp2/3 complex-mediated actin nucleation (7 genes)
positive regulation of histone h3-k9 acetylation (8 genes)
negative regulation of histone h4-k16 acetylation (4 genes)
negative regulation of nuclear-transcribed mrna catabolic process, nonsense-mediated decay (4 genes)
regulation of mirna metabolic process (8 genes)
positive regulation of mirna metabolic process (4 genes)
positive regulation of gene silencing by mirna (13 genes)
regulation of early endosome to late endosome transport (8 genes)
positive regulation of early endosome to late endosome transport (8 genes)
positive regulation of receptor catabolic process (4 genes)
negative regulation of stem cell proliferation (7 genes)
positive regulation of stem cell proliferation (21 genes)
regulation of sodium ion transmembrane transporter activity (17 genes)
negative regulation of sodium ion transmembrane transporter activity (8 genes)
positive regulation of sodium ion transmembrane transporter activity (8 genes)
regulation of genetic imprinting (7 genes)
negative regulation of interleukin-1-mediated signaling pathway (4 genes)
positive regulation of interleukin-5 secretion (6 genes)
positive regulation of interleukin-13 secretion (4 genes)
negative regulation of dendritic cell apoptotic process (4 genes)
negative regulation of motor neuron apoptotic process (7 genes)
negative regulation of type b pancreatic cell apoptotic process (6 genes)
positive regulation of type b pancreatic cell apoptotic process (6 genes)
negative regulation of transcription regulatory region dna binding (12 genes)
positive regulation of transcription regulatory region dna binding (16 genes)
negative regulation of cardiac muscle cell differentiation (7 genes)
positive regulation of cardiac muscle cell differentiation (11 genes)
regulation of stem cell differentiation (6 genes)
negative regulation of stem cell differentiation (18 genes)
positive regulation of stem cell differentiation (11 genes)
positive regulation of mesenchymal stem cell differentiation (4 genes)
negative regulation of peptidyl-lysine acetylation (6 genes)
negative regulation of cytoplasmic translation (8 genes)
positive regulation of cytoplasmic translation (5 genes)
regulation of cellular senescence (14 genes)
negative regulation of cellular senescence (19 genes)
positive regulation of cellular senescence (8 genes)
positive regulation of interleukin-6 secretion (27 genes)
negative regulation of double-strand break repair (4 genes)
positive regulation of double-strand break repair (12 genes)
regulation of autophagosome assembly (11 genes)
positive regulation of autophagosome assembly (11 genes)
positive regulation of synaptic vesicle clustering (8 genes)
regulation of bicellular tight junction assembly (10 genes)
negative regulation of anoikis (15 genes)
regulation of grooming behavior (6 genes)
positive regulation of ampa receptor activity (10 genes)
regulation of pro-b cell differentiation (4 genes)
positive regulation of behavioral fear response (7 genes)
epithelial cell proliferation involved in renal tubule morphogenesis (4 genes)
regulation of skeletal muscle cell differentiation (7 genes)
negative regulation of skeletal muscle cell differentiation (13 genes)
positive regulation of skeletal muscle cell differentiation (4 genes)
regulation of response to dna damage stimulus (10 genes)
positive regulation of response to dna damage stimulus (12 genes)
positive regulation of endothelial cell chemotaxis (11 genes)
regulation of double-strand break repair via nonhomologous end joining (5 genes)
negative regulation of double-strand break repair via nonhomologous end joining (4 genes)
positive regulation of double-strand break repair via nonhomologous end joining (10 genes)
positive regulation of integrin-mediated signaling pathway (9 genes)
negative regulation of mesenchymal cell apoptotic process (8 genes)
positive regulation of cysteine-type endopeptidase activity (10 genes)
regulation of endocytic recycling (4 genes)
positive regulation of endocytic recycling (5 genes)
regulation of rna biosynthetic process (5 genes)
positive regulation of histone h2b ubiquitination (4 genes)
negative regulation of atp biosynthetic process (4 genes)
positive regulation of atp biosynthetic process (11 genes)
positive regulation of interleukin-10 secretion (5 genes)
positive regulation of interleukin-12 secretion (4 genes)
negative regulation of dendritic cell differentiation (4 genes)
regulation of osteoclast development (4 genes)
negative regulation of osteoclast development (4 genes)
positive regulation of osteoclast development (4 genes)
positive regulation of vasculogenesis (9 genes)
regulation of neuron migration (19 genes)
negative regulation of neuron migration (13 genes)
positive regulation of neuron migration (19 genes)
negative regulation of apoptotic signaling pathway (32 genes)
positive regulation of apoptotic signaling pathway (40 genes)
regulation of extrinsic apoptotic signaling pathway (6 genes)
negative regulation of extrinsic apoptotic signaling pathway (42 genes)
positive regulation of extrinsic apoptotic signaling pathway (33 genes)
negative regulation of extrinsic apoptotic signaling pathway in absence of ligand (33 genes)
positive regulation of extrinsic apoptotic signaling pathway in absence of ligand (17 genes)
regulation of intrinsic apoptotic signaling pathway (6 genes)
negative regulation of intrinsic apoptotic signaling pathway (33 genes)
positive regulation of intrinsic apoptotic signaling pathway (33 genes)
regulation of store-operated calcium entry (14 genes)
negative regulation of cation channel activity (4 genes)
positive regulation of cation channel activity (13 genes)
regulation of semaphorin-plexin signaling pathway (6 genes)
negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway (6 genes)
positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway (5 genes)
phosphopyruvate hydratase complex (5 genes)
nucleotide-excision repair complex (6 genes)
histone deacetylase complex (35 genes)
rna polymerase i transcription factor complex (4 genes)
histone acetyltransferase complex (33 genes)
saga complex (13 genes)
pcaf complex (5 genes)
transcription factor tfiiib complex (6 genes)
transcription factor tfiiic complex (6 genes)
golgi cis cisterna (11 genes)
golgi trans cisterna (9 genes)
golgi membrane (148 genes)
exocyst (19 genes)
ubiquitin ligase complex (99 genes)
protein phosphatase type 2a complex (23 genes)
protein phosphatase type 1 complex (9 genes)
ribonuclease mrp complex (6 genes)
nuclear exosome (rnase complex) (16 genes)
cytoplasmic exosome (rnase complex) (12 genes)
exosome (rnase complex) (18 genes)
trna-intron endonuclease complex (4 genes)
vacuolar proton-transporting v-type atpase, v1 domain (4 genes)
nuclear chromosome (55 genes)
astral microtubule (4 genes)
pericentriolar material (21 genes)
commitment complex (7 genes)
mitochondrial proton-transporting atp synthase complex, catalytic core f(1) (6 genes)
mitochondrial proton-transporting atp synthase complex, coupling factor f(o) (11 genes)
cyclin-dependent protein kinase holoenzyme complex (30 genes)
storage vacuole (4 genes)
transcription export complex (13 genes)
tho complex (5 genes)
phagophore assembly site (24 genes)
ekc/keops complex (6 genes)
autophagosome membrane (15 genes)
core tfiih complex (7 genes)
mis12/mind type complex (4 genes)
tho complex part of transcription export complex (7 genes)
proteasome complex (62 genes)
glycosylphosphatidylinositol-n-acetylglucosaminyltransferase (gpi-gnt) complex (8 genes)
chromosome, centromeric region (150 genes)
kinetochore (127 genes)
condensed chromosome kinetochore (16 genes)
condensed nuclear chromosome kinetochore (10 genes)
condensed chromosome, centromeric region (9 genes)
condensed nuclear chromosome, centromeric region (19 genes)
chromosome, telomeric region (60 genes)
nuclear telomere cap complex (8 genes)
nuclear chromosome, telomeric region (77 genes)
chromatin (132 genes)
nucleosome (81 genes)
nuclear nucleosome (38 genes)
nuclear chromatin (236 genes)
euchromatin (10 genes)
heterochromatin (37 genes)
condensed chromosome (33 genes)
condensed nuclear chromosome (46 genes)
synaptonemal complex (65 genes)
condensin complex (8 genes)
lateral element (17 genes)
central element (8 genes)
x chromosome (5 genes)
origin recognition complex (6 genes)
swr1 complex (11 genes)
escrt i complex (10 genes)
escrt iii complex (9 genes)
spindle pole (82 genes)
equatorial microtubule organizing center (6 genes)
gamma-tubulin complex (15 genes)
p-body (73 genes)
garp complex (4 genes)
condensed chromosome outer kinetochore (10 genes)
condensed nuclear chromosome outer kinetochore (4 genes)
prp19 complex (15 genes)
mitochondrial sorting and assembly machinery complex (4 genes)
presequence translocase-associated import motor (5 genes)
voltage-gated sodium channel complex (14 genes)
outer dense fiber (9 genes)
microfibril (11 genes)
cornified envelope (55 genes)
fibrillar center (131 genes)
dense fibrillar component (6 genes)
granular component (7 genes)
acrosomal vesicle (111 genes)
male germ cell nucleus (24 genes)
female germ cell nucleus (4 genes)
stress fiber (78 genes)
ruffle (83 genes)
sex chromatin (6 genes)
barr body (5 genes)
xy body (22 genes)
photoreceptor outer segment (70 genes)
immunological synapse (36 genes)
phagocytic cup (18 genes)
photoreceptor inner segment (54 genes)
uropod (10 genes)
female pronucleus (9 genes)
male pronucleus (7 genes)
inner acrosomal membrane (4 genes)
acrosomal membrane (19 genes)
outer acrosomal membrane (5 genes)
podosome (24 genes)
semaphorin receptor complex (9 genes)
stereocilia ankle link (5 genes)
stereocilia ankle link complex (6 genes)
manchette (12 genes)
ribose phosphate diphosphokinase complex (6 genes)
zona pellucida receptor complex (13 genes)
cellular_component (3736 genes)
extracellular region (1656 genes)
fibrinogen complex (6 genes)
membrane attack complex (7 genes)
collagen trimer (76 genes)
collagen type iv trimer (7 genes)
collagen type v trimer (4 genes)
basement membrane (105 genes)
laminin-5 complex (4 genes)
interstitial matrix (17 genes)
extracellular space (1467 genes)
intracellular (736 genes)
cell (122 genes)
nucleus (5908 genes)
nuclear envelope (157 genes)
nuclear inner membrane (31 genes)
lamin filament (5 genes)
integral component of nuclear inner membrane (15 genes)
nuclear outer membrane (17 genes)
nuclear envelope lumen (13 genes)
annulate lamellae (5 genes)
nuclear pore (65 genes)
nuclear lamina (9 genes)
nucleoplasm (2019 genes)
nucleolar ribonuclease p complex (5 genes)
replication fork (25 genes)
alpha dna polymerase:primase complex (4 genes)
dna replication factor a complex (16 genes)
dna replication factor c complex (5 genes)
nuclear origin of replication recognition complex (8 genes)
dna-directed rna polymerase ii, core complex (15 genes)
dna-directed rna polymerase iii complex (18 genes)
transcription factor complex (277 genes)
transcription factor tfiid complex (38 genes)
ada2/gcn5/ada3 transcription activator complex (14 genes)
transcription factor tfiia complex (5 genes)
holo tfiih complex (11 genes)
chromatin silencing complex (6 genes)
anaphase-promoting complex (20 genes)
spliceosomal complex (150 genes)
u5 snrnp (16 genes)
u7 snrnp (5 genes)
u2-type spliceosomal complex (5 genes)
u1 snrnp (18 genes)
u2 snrnp (20 genes)
u4 snrnp (10 genes)
u6 snrnp (9 genes)
u12-type spliceosomal complex (26 genes)
chromosome (407 genes)
telomerase holoenzyme complex (20 genes)
nuclear euchromatin (30 genes)
nuclear heterochromatin (29 genes)
pericentric heterochromatin (16 genes)
perichromatin fibrils (6 genes)
nucleolus (695 genes)
small nucleolar ribonucleoprotein complex (12 genes)
dna-directed rna polymerase i complex (13 genes)
cytoplasm (6649 genes)
mitochondrion (1743 genes)
mitochondrial envelope (18 genes)
mitochondrial outer membrane (165 genes)
mitochondrial outer membrane translocase complex (10 genes)
mitochondrial inner membrane (408 genes)
mitochondrial inner membrane presequence translocase complex (7 genes)
mitochondrial respiratory chain (6 genes)
mitochondrial respiratory chain complex i (51 genes)
mitochondrial respiratory chain complex ii, succinate dehydrogenase complex (ubiquinone) (4 genes)
mitochondrial respiratory chain complex iii (12 genes)
mitochondrial respiratory chain complex iv (16 genes)
mitochondrial proton-transporting atp synthase complex (19 genes)
mitochondrial permeability transition pore complex (4 genes)
mitochondrial intermembrane space (70 genes)
mitochondrial matrix (124 genes)
mitochondrial ribosome (21 genes)
mitochondrial large ribosomal subunit (60 genes)
mitochondrial small ribosomal subunit (30 genes)
lysosome (377 genes)
lysosomal membrane (90 genes)
endosome (613 genes)
early endosome (215 genes)
late endosome (131 genes)
multivesicular body (33 genes)
vacuole (20 genes)
vacuolar membrane (20 genes)
autophagosome (52 genes)
peroxisome (136 genes)
peroxisomal membrane (40 genes)
integral component of peroxisomal membrane (19 genes)
peroxisomal matrix (19 genes)
endoplasmic reticulum (1449 genes)
sec61 translocon complex (6 genes)
signal recognition particle receptor complex (5 genes)
signal recognition particle, endoplasmic reticulum targeting (7 genes)
signal peptidase complex (7 genes)
endoplasmic reticulum lumen (51 genes)
endoplasmic reticulum membrane (296 genes)
smooth endoplasmic reticulum (31 genes)
rough endoplasmic reticulum (71 genes)
endoplasmic reticulum-golgi intermediate compartment (61 genes)
golgi apparatus (1259 genes)
golgi stack (27 genes)
golgi lumen (6 genes)
golgi medial cisterna (22 genes)
golgi-associated vesicle (15 genes)
cis-golgi network (43 genes)
trans-golgi network (188 genes)
lipid droplet (81 genes)
centrosome (490 genes)
centriole (109 genes)
microtubule organizing center (115 genes)
spindle (75 genes)
actomyosin contractile ring (8 genes)
polar microtubule (5 genes)
kinetochore microtubule (6 genes)
cytosol (3080 genes)
chaperonin-containing t-complex (10 genes)
hemoglobin complex (11 genes)
heterotrimeric g-protein complex (33 genes)
proteasome regulatory particle (10 genes)
proteasome core complex (20 genes)
ribosome (172 genes)
polysome (47 genes)
mrna cap binding complex (10 genes)
mrna cleavage and polyadenylation specificity factor complex (17 genes)
mrna cleavage factor complex (6 genes)
eukaryotic translation initiation factor 2 complex (6 genes)
eukaryotic translation initiation factor 2b complex (6 genes)
eukaryotic translation initiation factor 3 complex (15 genes)
cytoskeleton (1232 genes)
axonemal dynein complex (4 genes)
muscle myosin complex (6 genes)
troponin complex (8 genes)
striated muscle thin filament (21 genes)
cytoplasmic dynein complex (26 genes)
dynactin complex (9 genes)
kinesin complex (56 genes)
microtubule (349 genes)
microtubule associated complex (23 genes)
spindle microtubule (35 genes)
axonemal microtubule (5 genes)
cytoplasmic microtubule (60 genes)
intermediate filament (111 genes)
neurofilament (10 genes)
actin filament (86 genes)
arp2/3 protein complex (10 genes)
plasma membrane (4022 genes)
integral component of plasma membrane (1212 genes)
sodium:potassium-exchanging atpase complex (9 genes)
voltage-gated calcium channel complex (26 genes)
acetylcholine-gated channel complex (16 genes)
insulin receptor complex (5 genes)
caveola (83 genes)
microvillus (73 genes)
brush border (76 genes)
clathrin-coated pit (56 genes)
cell-cell junction (196 genes)
adherens junction (54 genes)
cell-cell adherens junction (91 genes)
zonula adherens (7 genes)
fascia adherens (13 genes)
gap junction (32 genes)
connexin complex (20 genes)
bicellular tight junction (121 genes)
focal adhesion (148 genes)
muscle tendon junction (5 genes)
cilium (318 genes)
axoneme (92 genes)
cell cortex (128 genes)
septin ring (14 genes)
phosphatidylinositol 3-kinase complex (22 genes)
camp-dependent protein kinase complex (8 genes)
calcineurin complex (5 genes)
phosphorylase kinase complex (5 genes)
rab-protein geranylgeranyltransferase complex (4 genes)
synaptic vesicle (158 genes)
transcription elongation factor complex (24 genes)
cyclin/cdk positive transcription elongation factor complex (9 genes)
voltage-gated potassium channel complex (75 genes)
spectrin (5 genes)
cop9 signalosome (36 genes)
oligosaccharyltransferase complex (9 genes)
gamma-tubulin ring complex (4 genes)
cohesin complex (7 genes)
inward rectifying potassium channel (4 genes)
f-actin capping protein complex (9 genes)
integrin complex (28 genes)
ionotropic glutamate receptor complex (16 genes)
ikappab kinase complex (7 genes)
proteasome regulatory particle, base subcomplex (12 genes)
proteasome regulatory particle, lid subcomplex (8 genes)
epsilon dna polymerase complex (4 genes)
external side of plasma membrane (372 genes)
cytoplasmic side of plasma membrane (58 genes)
basal plasma membrane (42 genes)
cell surface (653 genes)
endosome membrane (72 genes)
chromocenter (14 genes)
cytoplasmic stress granule (60 genes)
endomembrane system (40 genes)
vesicle membrane (28 genes)
er to golgi transport vesicle membrane (8 genes)
postsynaptic density (323 genes)
intercalated disc (57 genes)
spectrin-associated cytoskeleton (6 genes)
cajal body (35 genes)
actin cytoskeleton (198 genes)
microtubule cytoskeleton (172 genes)
large ribosomal subunit (14 genes)
small ribosomal subunit (17 genes)
dystrophin-associated glycoprotein complex (16 genes)
dystroglycan complex (5 genes)
sarcoglycan complex (6 genes)
membrane (6573 genes)
integral component of membrane (5425 genes)
inclusion body (19 genes)
aggresome (40 genes)
prefoldin complex (7 genes)
eukaryotic translation initiation factor 4f complex (5 genes)
eukaryotic 43s preinitiation complex (4 genes)
basolateral plasma membrane (200 genes)
apical plasma membrane (310 genes)
apicolateral plasma membrane (22 genes)
lateral plasma membrane (58 genes)
catenin complex (29 genes)
nuclear matrix (89 genes)
risc complex (10 genes)
myosin complex (47 genes)
myosin ii complex (7 genes)
unconventional myosin complex (5 genes)
vacuolar proton-transporting v-type atpase complex (15 genes)
swi/snf complex (17 genes)
sarcoplasm (8 genes)
sarcoplasmic reticulum (66 genes)
sin3 complex (13 genes)
nurd complex (18 genes)
nurf complex (7 genes)
dna-directed rna polymerase ii, holoenzyme (7 genes)
mediator complex (34 genes)
cdc73/paf1 complex (8 genes)
flotillin complex (10 genes)
ccaat-binding factor complex (5 genes)
nuclear body (289 genes)
pml body (72 genes)
nuclear speck (311 genes)
transcriptional repressor complex (57 genes)
serine c-palmitoyltransferase complex (5 genes)
intracellular cyclic nucleotide activated cation channel complex (4 genes)
aminoacyl-trna synthetase multienzyme complex (13 genes)
golgi transport complex (11 genes)
nmda selective glutamate receptor complex (14 genes)
scf ubiquitin ligase complex (60 genes)
proteasome core complex, alpha-subunit complex (8 genes)
b cell receptor complex (4 genes)
extrinsic component of plasma membrane (36 genes)
extrinsic component of membrane (61 genes)
symbiont-containing vacuole membrane (9 genes)
ciliary pocket membrane (4 genes)
proteasome accessory complex (17 genes)
cytosolic large ribosomal subunit (61 genes)
cytosolic ribosome (16 genes)
cytosolic small ribosomal subunit (44 genes)
trapp complex (12 genes)
ccr4-not complex (17 genes)
ccr4-not core complex (5 genes)
myofibril (41 genes)
sarcomere (40 genes)
z disc (125 genes)
lamellipodium (156 genes)
cell junction (806 genes)
hemidesmosome (10 genes)
desmosome (25 genes)
mitochondrial crista (12 genes)
membrane coat (28 genes)
clathrin coat (7 genes)
ap-1 adaptor complex (5 genes)
ap-2 adaptor complex (14 genes)
ap-3 adaptor complex (12 genes)
ap-4 adaptor complex (5 genes)
clathrin vesicle coat (4 genes)
copi vesicle coat (14 genes)
copii vesicle coat (14 genes)
clathrin coat of trans-golgi network vesicle (4 genes)
clathrin adaptor complex (17 genes)
clathrin coat of coated pit (8 genes)
transport vesicle (43 genes)
copii-coated er to golgi transport vesicle (36 genes)
coated vesicle (4 genes)
clathrin-coated vesicle (49 genes)
copi-coated vesicle (12 genes)
endocytic vesicle (58 genes)
trans-golgi network transport vesicle (16 genes)
secretory granule (143 genes)
integral component of golgi membrane (49 genes)
filopodium (61 genes)
integral component of endoplasmic reticulum membrane (107 genes)
integral component of synaptic vesicle membrane (45 genes)
dynein complex (42 genes)
protein phosphatase 4 complex (5 genes)
junctional membrane complex (8 genes)
t-tubule (63 genes)
axon (423 genes)
dendrite (510 genes)
growth cone (155 genes)
actin cap (4 genes)
smooth muscle contractile fiber (5 genes)
midbody (132 genes)
small nuclear ribonucleoprotein complex (8 genes)
cytoplasmic vesicle membrane (42 genes)
golgi-associated vesicle membrane (10 genes)
clathrin-coated vesicle membrane (4 genes)
secretory granule membrane (29 genes)
phagocytic vesicle membrane (5 genes)
synaptic vesicle membrane (56 genes)
axolemma (21 genes)
multimeric ribonuclease p complex (8 genes)
90s preribosome (8 genes)
preribosome, large subunit precursor (22 genes)
preribosome, small subunit precursor (9 genes)
cortical cytoskeleton (31 genes)
cortical actin cytoskeleton (50 genes)
rough endoplasmic reticulum membrane (4 genes)
smooth endoplasmic reticulum membrane (5 genes)
mre11 complex (6 genes)
beta-catenin destruction complex (10 genes)
vcb complex (6 genes)
meiotic cohesin complex (7 genes)
checkpoint clamp complex (6 genes)
hops complex (11 genes)
retromer complex (21 genes)
staga complex (13 genes)
smc5-smc6 complex (7 genes)
intraciliary transport particle a (7 genes)
intraciliary transport particle b (22 genes)
ino80 complex (15 genes)
extracellular matrix (280 genes)
interphase microtubule organizing center (4 genes)
dense core granule (18 genes)
nuclear pore outer ring (11 genes)
bloc-1 complex (15 genes)
organelle membrane (4 genes)
platelet alpha granule (18 genes)
septin complex (14 genes)
pseudopodium (12 genes)
snare complex (48 genes)
scar complex (8 genes)
intrinsic component of membrane (16 genes)
anchored component of membrane (148 genes)
intrinsic component of plasma membrane (32 genes)
intrinsic component of endoplasmic reticulum membrane (9 genes)
extrinsic component of external side of plasma membrane (8 genes)
extrinsic component of cytoplasmic side of plasma membrane (63 genes)
cell leading edge (61 genes)
cell projection membrane (13 genes)
leading edge membrane (8 genes)
lamellipodium membrane (9 genes)
ndc80 complex (4 genes)
death-inducing signaling complex (6 genes)
cd95 death-inducing signaling complex (4 genes)
replication fork protection complex (8 genes)
integral component of organelle membrane (10 genes)
integral component of mitochondrial inner membrane (30 genes)
integral component of mitochondrial outer membrane (22 genes)
extrinsic component of endosome membrane (9 genes)
extrinsic component of mitochondrial inner membrane (8 genes)
extrinsic component of mitochondrial outer membrane (5 genes)
anchored component of external side of plasma membrane (22 genes)
ctf18 rfc-like complex (8 genes)
cytoplasmic vesicle (738 genes)
nata complex (5 genes)
box c/d snornp complex (6 genes)
box h/aca snornp complex (4 genes)
m band (22 genes)
cullin-ring ubiquitin ligase complex (13 genes)
cul2-ring ubiquitin ligase complex (14 genes)
cul3-ring ubiquitin ligase complex (35 genes)
cul4a-ring e3 ubiquitin ligase complex (10 genes)
cul4b-ring e3 ubiquitin ligase complex (4 genes)
cul5-ring ubiquitin ligase complex (5 genes)
cul7-ring ubiquitin ligase complex (5 genes)
motile cilium (132 genes)
pcg protein complex (28 genes)
brush border membrane (77 genes)
filopodium membrane (18 genes)
microvillus membrane (19 genes)
nucleotide-activated protein kinase complex (14 genes)
neuromuscular junction (98 genes)
cytosolic proteasome complex (8 genes)
spindle pole centrosome (19 genes)
nuclear pericentric heterochromatin (9 genes)
a band (22 genes)
i band (22 genes)
haptoglobin-hemoglobin complex (12 genes)
early endosome membrane (43 genes)
late endosome membrane (37 genes)
torc1 complex (7 genes)
torc2 complex (12 genes)
filamentous actin (32 genes)
nuclear membrane (200 genes)
mitochondrial membrane (88 genes)
nuclear lumen (4 genes)
vesicle (143 genes)
golgi cisterna (14 genes)
early phagosome (8 genes)
nelf complex (4 genes)
integrator complex (18 genes)
small-subunit processome (37 genes)
guanyl-nucleotide exchange factor complex (11 genes)
chromosome passenger complex (5 genes)
cleavage furrow (41 genes)
septin filament array (14 genes)
asymmetric synapse (21 genes)
ampa glutamate receptor complex (32 genes)
mismatch repair complex (10 genes)
mutlalpha complex (6 genes)
photoreceptor connecting cilium (44 genes)
stereocilium (36 genes)
stereocilium bundle (15 genes)
stereocilium tip (19 genes)
actin filament bundle (10 genes)
filopodium tip (18 genes)
cuticular plate (7 genes)
cortical endoplasmic reticulum (7 genes)
golgi cisterna membrane (14 genes)
growth cone membrane (7 genes)
multivesicular body membrane (6 genes)
ruffle membrane (70 genes)
trans-golgi network membrane (31 genes)
dendrite membrane (41 genes)
dendritic spine membrane (11 genes)
integral component of mitochondrial membrane (9 genes)
insulin-responsive compartment (8 genes)
piccolo nua4 histone acetyltransferase complex (4 genes)
ell-eaf complex (5 genes)
smn complex (11 genes)
neuronal cell body membrane (35 genes)
plasma membrane bounded cell projection cytoplasm (4 genes)
dendrite cytoplasm (30 genes)
myosin filament (12 genes)
kainate selective glutamate receptor complex (6 genes)
macromolecular complex (811 genes)
protein-dna complex (52 genes)
paranodal junction (5 genes)
sarcoplasmic reticulum membrane (21 genes)
rad51b-rad51c-rad51d-xrcc2 complex (4 genes)
weibel-palade body (5 genes)
endoplasmic reticulum-golgi intermediate compartment membrane (8 genes)
melanosome membrane (6 genes)
proton-transporting two-sector atpase complex, proton-transporting domain (5 genes)
proton-transporting v-type atpase, v0 domain (9 genes)
proton-transporting v-type atpase, v1 domain (7 genes)
corvet complex (6 genes)
node of ranvier (17 genes)
internode region of axon (4 genes)
paranode region of axon (17 genes)
transcription factor tftc complex (15 genes)
chromatoid body (15 genes)
hulc complex (4 genes)
rdna heterochromatin (5 genes)
elongator holoenzyme complex (6 genes)
phagophore assembly site membrane (12 genes)
vcp-npl4-ufd1 aaa atpase complex (8 genes)
phosphatidylinositol 3-kinase complex, class iii, type i (4 genes)
phosphatidylinositol 3-kinase complex, class iii, type ii (4 genes)
very-low-density lipoprotein particle (18 genes)
low-density lipoprotein particle (13 genes)
intermediate-density lipoprotein particle (5 genes)
high-density lipoprotein particle (20 genes)
spherical high-density lipoprotein particle (7 genes)
pwp2p-containing subcomplex of 90s preribosome (6 genes)
nuclear periphery (22 genes)
ego complex (5 genes)
centriolar satellite (31 genes)
bbsome (8 genes)
gid complex (4 genes)
endoplasmic reticulum chaperone complex (13 genes)
ion channel complex (8 genes)
cation channel complex (10 genes)
calcium channel complex (18 genes)
potassium channel complex (12 genes)
sodium channel complex (9 genes)
chloride channel complex (46 genes)
methylosome (10 genes)
picln-sm protein complex (5 genes)
smn-sm protein complex (15 genes)
aryl hydrocarbon receptor complex (4 genes)
nuclear meiotic cohesin complex (5 genes)
linc complex (11 genes)
phosphatidylinositol 3-kinase complex, class iii (6 genes)
interchromatin granule (4 genes)
micro-ribonucleoprotein complex (116 genes)
histone methyltransferase complex (47 genes)
esc/e(z) complex (16 genes)
prc1 complex (15 genes)
exon-exon junction complex (21 genes)
ciliary rootlet (13 genes)
nua4 histone acetyltransferase complex (20 genes)
transcriptionally active chromatin (26 genes)
spots complex (4 genes)
histone locus body (4 genes)
microtubule plus-end (21 genes)
pr-dub complex (4 genes)
cd40 receptor complex (12 genes)
mon1-ccz1 complex (4 genes)
sperm fibrous sheath (14 genes)
myelin sheath abaxonal region (9 genes)
myelin sheath adaxonal region (6 genes)
ribonucleoprotein granule (15 genes)
site of double-strand break (47 genes)
alphav-beta3 integrin-igf-1-igf1r complex (4 genes)
ciliary transition zone (34 genes)
transcription factor ap-1 complex (6 genes)
senescence-associated heterochromatin focus (4 genes)
mks complex (14 genes)
slit diaphragm (9 genes)
ciliary basal body (136 genes)
sperm flagellum (56 genes)
catsper complex (13 genes)
inner dynein arm (11 genes)
outer dynein arm (10 genes)
rna n6-methyladenosine methyltransferase complex (8 genes)
microtubule minus-end (7 genes)
keratohyalin granule (4 genes)
cytoplasmic ribonucleoprotein granule (66 genes)
somatodendritic compartment (19 genes)
derlin-1 retrotranslocation complex (10 genes)
t cell receptor complex (13 genes)
alpha-beta t cell receptor complex (5 genes)
paraspeckles (7 genes)
sarcolemma (119 genes)
nuclear inclusion body (12 genes)
extrinsic component of endoplasmic reticulum membrane (6 genes)
melanosome (29 genes)
mcm complex (9 genes)
insulin-like growth factor ternary complex (4 genes)
specific granule (13 genes)
azurophil granule (9 genes)
germinal vesicle (4 genes)
glycogen granule (9 genes)
zymogen granule (12 genes)
zymogen granule membrane (14 genes)
mhc class i protein complex (11 genes)
mhc class ii protein complex (9 genes)
photoreceptor outer segment membrane (13 genes)
chylomicron (12 genes)
mast cell granule (7 genes)
actomyosin (12 genes)
mitochondrial nucleoid (47 genes)
mitochondrial intermembrane space protein transporter complex (5 genes)
mitochondrial inner membrane protein insertion complex (6 genes)
presynaptic membrane (80 genes)
gpi-anchor transamidase complex (5 genes)
polysomal ribosome (30 genes)
mhc class i peptide loading complex (5 genes)
cell projection (916 genes)
neuron projection (522 genes)
nadph oxidase complex (10 genes)
neuronal cell body (605 genes)
costamere (19 genes)
synaptic cleft (19 genes)
acrosomal matrix (6 genes)
p granule (11 genes)
axon initial segment (21 genes)
terminal bouton (83 genes)
varicosity (16 genes)
dendritic spine (176 genes)
dendritic shaft (68 genes)
lysosomal lumen (5 genes)
axon hillock (9 genes)
perikaryon (137 genes)
myelin sheath (198 genes)
compact myelin (5 genes)
lateral loop (7 genes)
schmidt-lanterman incisure (15 genes)
membrane-bounded organelle (5 genes)
intracellular organelle (15 genes)
intracellular membrane-bounded organelle (869 genes)
receptor complex (212 genes)
fanconi anaemia nuclear complex (14 genes)
laminin complex (4 genes)
contractile fiber (20 genes)
apical junction complex (23 genes)
nuclear replication fork (11 genes)
delta dna polymerase complex (4 genes)
axon terminus (88 genes)
cytolytic granule (5 genes)
spanning component of plasma membrane (9 genes)
juxtaparanode region of axon (10 genes)
organelle membrane contact site (6 genes)
er-mitochondrion membrane contact site (12 genes)
cell-cell contact zone (20 genes)
dendrite terminus (8 genes)
dendritic growth cone (12 genes)
axonal growth cone (42 genes)
cell body (112 genes)
c-fiber (4 genes)
cerebellar mossy fiber (10 genes)
main axon (6 genes)
calyx of held (29 genes)
neuron projection terminus (24 genes)
dendritic branch (4 genes)
axonal spine (4 genes)
neuron spine (11 genes)
cone cell pedicle (5 genes)
endoplasmic reticulum quality control compartment (7 genes)
dendritic spine neck (7 genes)
dendritic spine head (15 genes)
macropinosome (6 genes)
supraspliceosomal complex (4 genes)
nuclear pore central transport channel (8 genes)
nuclear pore nuclear basket (6 genes)
mll3/4 complex (10 genes)
autolysosome (13 genes)
nuclear transcription factor complex (25 genes)
plasma membrane raft (26 genes)
keratin filament (41 genes)
type iii intermediate filament (5 genes)
intermediate filament cytoskeleton (53 genes)
pronucleus (15 genes)
membrane raft (273 genes)
intercellular bridge (56 genes)
apical part of cell (121 genes)
basal part of cell (19 genes)
apical cortex (9 genes)
basal cortex (7 genes)
synapse (699 genes)
postsynaptic membrane (263 genes)
oxoglutarate dehydrogenase complex (6 genes)
pyruvate dehydrogenase complex (8 genes)
proton-transporting atp synthase complex (4 genes)
proton-transporting atp synthase complex, catalytic core f(1) (6 genes)
proton-transporting atp synthase complex, coupling factor f(o) (12 genes)
respiratory chain complex iv (9 genes)
clathrin-coated endocytic vesicle (13 genes)
phagocytic vesicle (74 genes)
u4/u6 x u5 tri-snrnp complex (30 genes)
intercellular canaliculus (10 genes)
anchored component of plasma membrane (44 genes)
lipopolysaccharide receptor complex (5 genes)
pore complex (8 genes)
activin receptor complex (7 genes)
set1c/compass complex (13 genes)
rough endoplasmic reticulum lumen (6 genes)
perinuclear region of cytoplasm (622 genes)
holliday junction resolvase complex (5 genes)
signal recognition particle (8 genes)
presynaptic active zone (37 genes)
presynaptic active zone membrane (17 genes)
cytoskeleton of presynaptic active zone (7 genes)
spindle midzone (24 genes)
cell tip (7 genes)
recycling endosome (105 genes)
recycling endosome membrane (42 genes)
excitatory synapse (39 genes)
inhibitory synapse (25 genes)
kinocilium (8 genes)
ciliary membrane (33 genes)
asap complex (5 genes)
micos complex (8 genes)
mitotic spindle astral microtubule (5 genes)
tricellular tight junction (4 genes)
gator2 complex (11 genes)
inflammasome complex (14 genes)
sperm head (9 genes)
leading edge of lamellipodium (4 genes)
synaptobrevin 2-snap-25-syntaxin-1a-complexin i complex (4 genes)
synaptobrevin 2-snap-25-syntaxin-1a-complexin ii complex (4 genes)
synaptobrevin 2-snap-25-syntaxin-1a complex (5 genes)
extracellular exosome (84 genes)
ciliary neurotrophic factor receptor complex (4 genes)
occluding junction (5 genes)
shelterin complex (7 genes)
exocytic vesicle (29 genes)
nonhomologous end joining complex (8 genes)
elongin complex (4 genes)
saga-type complex (6 genes)
respiratory chain (60 genes)
ercc4-ercc1 complex (5 genes)
brca1-a complex (8 genes)
pebow complex (6 genes)
brisc complex (6 genes)
risc-loading complex (11 genes)
swi/snf superfamily-type complex (17 genes)
haus complex (9 genes)
fhf complex (5 genes)
pre-snornp complex (8 genes)
gamma-secretase complex (7 genes)
moz/morf histone acetyltransferase complex (7 genes)
tertiary granule (4 genes)
sin3-type complex (7 genes)
core mediator complex (8 genes)
cell body fiber (12 genes)
soss complex (4 genes)
crd-mediated mrna stability complex (7 genes)
contractile ring (6 genes)
endoplasmic reticulum exit site (25 genes)
u2-type prespliceosome (14 genes)
u2-type catalytic step 1 spliceosome (5 genes)
u2-type catalytic step 2 spliceosome (28 genes)
catalytic step 2 spliceosome (80 genes)
post-mrna release spliceosomal complex (7 genes)
smad protein complex (4 genes)
wash complex (10 genes)
histone pre-mrna 3'end processing complex (6 genes)
mll1 complex (30 genes)
invadopodium (10 genes)
invadopodium membrane (5 genes)
clathrin complex (9 genes)
integral component of cytoplasmic side of endoplasmic reticulum membrane (6 genes)
eukaryotic translation initiation factor 3 complex, eif3m (8 genes)
pi-body (7 genes)
pip-body (5 genes)
integral component of lumenal side of endoplasmic reticulum membrane (6 genes)
npbaf complex (12 genes)
nbaf complex (15 genes)
neuronal ribonucleoprotein granule (6 genes)
endoplasmic reticulum tubular network (14 genes)
lubac complex (5 genes)
mmxd complex (5 genes)
cell periphery (73 genes)
elastic fiber (4 genes)
ragulator complex (6 genes)
ptw/pp1 phosphatase complex (7 genes)
msl complex (5 genes)
er membrane protein complex (11 genes)
ipaf inflammasome complex (5 genes)
nlrp1 inflammasome complex (6 genes)
nlrp3 inflammasome complex (5 genes)
blood microparticle (4 genes)
trna-splicing ligase complex (8 genes)
mitotic spindle (73 genes)
meiotic spindle (12 genes)
cul4-ring e3 ubiquitin ligase complex (27 genes)
u2af (5 genes)
flemming body (15 genes)
rna polymerase ii transcription repressor complex (4 genes)
rna polymerase ii transcription factor complex (76 genes)
central region of growth cone (6 genes)
site of dna damage (12 genes)
mpp7-dlg1-lin7 complex (5 genes)
perinuclear endoplasmic reticulum (20 genes)
traf2-gstp1 complex (5 genes)
cntfr-clcf1 complex (4 genes)
synaptic membrane (49 genes)
bcl-2 family protein complex (5 genes)
nuclear stress granule (5 genes)
tetraspanin-enriched microdomain (9 genes)
alveolar lamellar body (9 genes)
epidermal lamellar body (4 genes)
sperm connecting piece (4 genes)
sperm midpiece (28 genes)
sperm annulus (7 genes)
sperm principal piece (30 genes)
r2tp complex (7 genes)
ripoptosome (6 genes)
cia complex (5 genes)
glial cell projection (11 genes)
neurofibrillary tangle (4 genes)
tubular endosome (5 genes)
microtubule bundle (6 genes)
mitotic spindle pole (30 genes)
dense body (5 genes)
apical dendrite (26 genes)
basal dendrite (8 genes)
sorting endosome (5 genes)
spine apparatus (4 genes)
astrocyte projection (11 genes)
gait complex (4 genes)
neuron part (11 genes)
ribbon synapse (10 genes)
gemini of coiled bodies (7 genes)
cardiac myofibril (9 genes)
sperm plasma membrane (5 genes)
ciliary transition fiber (10 genes)
ciliary tip (11 genes)
ciliary base (35 genes)
transcriptional preinitiation complex (7 genes)
integral component of autophagosome membrane (4 genes)
intracellular vesicle (7 genes)
9+2 motile cilium (8 genes)
non-motile cilium (36 genes)
deuterosome (6 genes)
cytoplasmic side of lysosomal membrane (4 genes)
protein complex involved in cell adhesion (5 genes)
hippocampal mossy fiber to ca3 synapse (49 genes)
presynapse (100 genes)
postsynapse (118 genes)
plasma membrane protein complex (9 genes)
endoplasmic reticulum tubular network membrane (5 genes)
presynaptic active zone cytoplasmic component (23 genes)
presynaptic endocytic zone (5 genes)
presynaptic endocytic zone membrane (7 genes)
postsynaptic recycling endosome (4 genes)
postsynaptic density membrane (31 genes)
postsynaptic endosome (4 genes)
extrinsic component of synaptic vesicle membrane (10 genes)
postsynaptic actin cytoskeleton (14 genes)
extrinsic component of presynaptic membrane (5 genes)
extrinsic component of postsynaptic membrane (7 genes)
glutamatergic synapse (511 genes)
gaba-ergic synapse (100 genes)
neuronal dense core vesicle (6 genes)
anchored component of synaptic vesicle membrane (20 genes)
integral component of postsynaptic membrane (82 genes)
integral component of presynaptic membrane (92 genes)
integral component of presynaptic active zone membrane (25 genes)
integral component of postsynaptic specialization membrane (37 genes)
integral component of postsynaptic density membrane (75 genes)
borc complex (8 genes)
secretory vesicle (7 genes)
presynaptic cytosol (25 genes)
postsynaptic cytosol (27 genes)
postsynaptic specialization (5 genes)
postsynaptic specialization membrane (4 genes)
cell cortex region (13 genes)
apical plasma membrane urothelial plaque (4 genes)
plasma membrane bounded cell projection part (4 genes)
centriolar subdistal appendage (9 genes)
kicstor complex (5 genes)
dendritic microtubule (4 genes)
catalytic complex (5 genes)
gaba receptor complex (5 genes)
gaba-a receptor complex (19 genes)
extracellular vesicle (4 genes)
axon cytoplasm (15 genes)
mitotic spindle midzone (15 genes)
periciliary membrane compartment (6 genes)
spermatoproteasome complex (5 genes)
messenger ribonucleoprotein complex (14 genes)
gator1 complex (6 genes)
gtr1-gtr2 gtpase complex (4 genes)
transferase complex (4 genes)
uniplex complex (5 genes)
atg1/ulk1 kinase complex (8 genes)
terminal web (4 genes)
dna repair complex (6 genes)
3m complex (4 genes)
peroxisomal importomer complex (6 genes)
l-type voltage-gated calcium channel complex (14 genes)
mitotic spindle microtubule (4 genes)
ire1-traf2-ask1 complex (4 genes)
proximal dendrite (8 genes)
ush2 complex (4 genes)
hfe-transferrin receptor complex (6 genes)
lsm1-7-pat1 complex (5 genes)
earp complex (4 genes)
proximal neuron projection (5 genes)
growth cone filopodium (4 genes)
wnt-frizzled-lrp5/6 complex (4 genes)
ribonucleoprotein complex (163 genes)
beta-catenin-tcf complex (10 genes)
wnt signalosome (9 genes)
sperm head plasma membrane (7 genes)
single-stranded dna endodeoxyribonuclease activity (9 genes)
mannosyltransferase activity (14 genes)
trna binding (61 genes)
fatty-acyl-coa binding (25 genes)
l-ornithine transmembrane transporter activity (7 genes)
microfilament motor activity (15 genes)
snare binding (69 genes)
phosphorelay sensor kinase activity (5 genes)
nucleotide binding (1716 genes)
3'-5'-exoribonuclease activity (21 genes)
rdna binding (4 genes)
trna-intron endonuclease activity (4 genes)
3-keto sterol reductase activity (4 genes)
magnesium ion binding (198 genes)
ferric-chelate reductase activity (4 genes)
endopolyphosphatase activity (5 genes)
rna cap binding (6 genes)
rna 7-methylguanosine cap binding (9 genes)
four-way junction dna binding (15 genes)
y-form dna binding (6 genes)
bubble dna binding (5 genes)
adenyl-nucleotide exchange factor activity (9 genes)
inositol hexakisphosphate binding (4 genes)
inositol hexakisphosphate kinase activity (6 genes)
inositol hexakisphosphate 5-kinase activity (6 genes)
translation repressor activity, nucleic acid binding (13 genes)
transcription regulatory region sequence-specific dna binding (84 genes)
rna polymerase ii regulatory region sequence-specific dna binding (306 genes)
rna polymerase ii proximal promoter sequence-specific dna binding (511 genes)
rna polymerase ii core promoter sequence-specific dna binding (25 genes)
rna polymerase ii distal enhancer sequence-specific dna binding (94 genes)
rna polymerase ii transcription factor activity, sequence-specific dna binding (245 genes)
transcription factor activity, rna polymerase ii proximal promoter sequence-specific dna binding (21 genes)
transcription factor activity, rna polymerase ii core promoter sequence-specific dna binding (14 genes)
proximal promoter sequence-specific dna binding (41 genes)
rna polymerase ii core binding (31 genes)
transcription factor activity, core rna polymerase iii binding (7 genes)
rna polymerase ii regulatory region dna binding (14 genes)
rna polymerase iii regulatory region dna binding (4 genes)
rna polymerase iii type 3 promoter dna binding (4 genes)
core promoter sequence-specific dna binding (6 genes)
core promoter binding (14 genes)
rna polymerase i activity (12 genes)
rna polymerase ii activity (9 genes)
rna polymerase iii activity (16 genes)
regulatory region rna binding (5 genes)
transcription factor activity, rna polymerase ii core promoter sequence-specific binding involved in preinitiation complex assembly (10 genes)
transcriptional activator activity, rna polymerase ii proximal promoter sequence-specific dna binding (328 genes)
transcriptional repressor activity, rna polymerase ii proximal promoter sequence-specific dna binding (187 genes)
rna polymerase ii transcription factor binding (62 genes)
tfiib-class transcription factor binding (4 genes)
tfiid-class transcription factor binding (11 genes)
tfiih-class transcription factor binding (5 genes)
rna polymerase ii activating transcription factor binding (45 genes)
rna polymerase ii repressing transcription factor binding (39 genes)
enhancer sequence-specific dna binding (30 genes)
rna polymerase ii intronic transcription regulatory region sequence-specific dna binding (11 genes)
rna polymerase i core element sequence-specific dna binding (8 genes)
transcriptional activator activity, rna polymerase ii distal enhancer sequence-specific dna binding (42 genes)
transcriptional repressor activity, rna polymerase ii distal enhancer sequence-specific binding (10 genes)
transcription cofactor binding (10 genes)
transcription corepressor binding (17 genes)
transcription coactivator binding (18 genes)
transcriptional repressor activity, rna polymerase ii transcription regulatory region sequence-specific dna binding (95 genes)
transcriptional activator activity, rna polymerase ii transcription regulatory region sequence-specific dna binding (166 genes)
opioid peptide activity (5 genes)
n-acetylglucosamine 6-o-sulfotransferase activity (6 genes)
lipopolysaccharide binding (27 genes)
n-acetylgalactosamine 4-o-sulfotransferase activity (5 genes)
amyloid-beta binding (55 genes)
dopamine neurotransmitter receptor activity, coupled via gi/go (9 genes)
trace-amine receptor activity (15 genes)
peptide yy receptor activity (5 genes)
pancreatic polypeptide receptor activity (4 genes)
adrenomedullin receptor activity (4 genes)
g-protein coupled adenosine receptor activity (6 genes)
purinergic nucleotide receptor activity (8 genes)
virus receptor activity (19 genes)
calcitonin gene-related peptide receptor activity (4 genes)
group iii metabotropic glutamate receptor activity (4 genes)
peptide receptor activity (8 genes)
g-protein coupled receptor binding (67 genes)
alpha-n-acetylgalactosaminide alpha-2,6-sialyltransferase activity (5 genes)
atpase activator activity (25 genes)
l-amino-acid oxidase activity (4 genes)
2'-5'-oligoadenylate synthetase activity (6 genes)
retinal dehydrogenase activity (8 genes)
phosphotyrosine residue binding (45 genes)
phosphatidylserine binding (58 genes)
complement binding (9 genes)
complement component c1q binding (8 genes)
lipopolysaccharide receptor activity (4 genes)
nucleoside binding (5 genes)
purine nucleoside binding (5 genes)
g-protein alpha-subunit binding (33 genes)
fibronectin binding (31 genes)
retinoic acid binding (15 genes)
protease binding (121 genes)
p53 binding (70 genes)
opsin binding (4 genes)
utp binding (4 genes)
g-quadruplex rna binding (10 genes)
aminoacyl-trna editing activity (12 genes)
dystroglycan binding (11 genes)
ceramide phosphoethanolamine synthase activity (4 genes)
molecular_function (4920 genes)
nucleic acid binding (845 genes)
dna binding (1753 genes)
dna helicase activity (15 genes)
at dna binding (10 genes)
chromatin binding (480 genes)
damaged dna binding (52 genes)
dna replication origin binding (16 genes)
dna clamp loader activity (9 genes)
double-stranded dna binding (157 genes)
double-stranded telomeric dna binding (9 genes)
single-stranded dna binding (93 genes)
dna binding transcription factor activity (735 genes)
transcription factor activity, rna polymerase ii distal enhancer sequence-specific binding (74 genes)
steroid hormone receptor activity (50 genes)
retinoic acid receptor activity (4 genes)
transcription cofactor activity (67 genes)
transcription coactivator activity (220 genes)
transcription corepressor activity (187 genes)
telomerase activity (7 genes)
rna binding (784 genes)
rna helicase activity (4 genes)
double-stranded rna binding (81 genes)
double-stranded rna adenosine deaminase activity (6 genes)
single-stranded rna binding (46 genes)
mrna binding (174 genes)
mrna 3'-utr binding (81 genes)
structural constituent of ribosome (151 genes)
translation initiation factor activity (47 genes)
translation elongation factor activity (18 genes)
translation release factor activity (7 genes)
peptidyl-prolyl cis-trans isomerase activity (34 genes)
protein disulfide isomerase activity (16 genes)
motor activity (70 genes)
microtubule motor activity (67 genes)
actin binding (355 genes)
actin monomer binding (26 genes)
lysozyme activity (10 genes)
protein-glutamine gamma-glutamyltransferase activity (8 genes)
antigen binding (14 genes)
catalytic activity (469 genes)
alpha-n-acetylneuraminate alpha-2,8-sialyltransferase activity (5 genes)
beta-n-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity (5 genes)
beta-galactoside (cmp) alpha-2,3-sialyltransferase activity (4 genes)
1-acylglycerol-3-phosphate o-acyltransferase activity (11 genes)
2-acylglycerol o-acyltransferase activity (4 genes)
1-alkyl-2-acetylglycerophosphocholine esterase activity (5 genes)
3-beta-hydroxy-delta5-steroid dehydrogenase activity (10 genes)
3-hydroxyacyl-coa dehydrogenase activity (8 genes)
6-phosphofructo-2-kinase activity (4 genes)
dna-directed dna polymerase activity (25 genes)
dna-directed 5'-3' rna polymerase activity (36 genes)
dna-(apurinic or apyrimidinic site) lyase activity (12 genes)
dna topoisomerase activity (6 genes)
dna topoisomerase type i activity (4 genes)
dna topoisomerase type ii (atp-hydrolyzing) activity (4 genes)
gpi-anchor transamidase activity (5 genes)
gtpase activity (294 genes)
n-acetyllactosamine synthase activity (5 genes)
nad+ adp-ribosyltransferase activity (23 genes)
nad+ kinase activity (13 genes)
nadh dehydrogenase activity (17 genes)
nad(p)+-protein-arginine adp-ribosyltransferase activity (6 genes)
nadph-hemoprotein reductase activity (9 genes)
acetyl-coa c-acetyltransferase activity (5 genes)
acetyl-coa c-acyltransferase activity (9 genes)
acid phosphatase activity (8 genes)
acyl-coa dehydrogenase activity (9 genes)
acyl-coa ligase activity (5 genes)
acyl-coa oxidase activity (4 genes)
adenosine deaminase activity (8 genes)
atp-dependent dna helicase activity (25 genes)
atp-dependent rna helicase activity (20 genes)
phospholipid-translocating atpase activity (21 genes)
adenylate cyclase activity (12 genes)
adenylate kinase activity (9 genes)
alcohol dehydrogenase (nad) activity (6 genes)
alcohol sulfotransferase activity (7 genes)
3-chloroallyl aldehyde dehydrogenase activity (9 genes)
aldehyde dehydrogenase (nad) activity (16 genes)
aldehyde dehydrogenase [nad(p)+] activity (4 genes)
aldehyde oxidase activity (5 genes)
alditol:nadp+ 1-oxidoreductase activity (18 genes)
aldo-keto reductase (nadp) activity (8 genes)
alkaline phosphatase activity (4 genes)
aminoacyl-trna hydrolase activity (4 genes)
aminoacylase activity (4 genes)
arachidonate 12-lipoxygenase activity (4 genes)
arylamine n-acetyltransferase activity (4 genes)
aryl sulfotransferase activity (4 genes)
arylesterase activity (6 genes)
arylsulfatase activity (4 genes)
carbonate dehydratase activity (15 genes)
carbonyl reductase (nadph) activity (5 genes)
carnitine o-palmitoyltransferase activity (4 genes)
catalase activity (4 genes)
creatine kinase activity (5 genes)
cyclic-nucleotide phosphodiesterase activity (5 genes)
3',5'-cyclic-nucleotide phosphodiesterase activity (25 genes)
3',5'-cyclic-amp phosphodiesterase activity (14 genes)
cgmp-stimulated cyclic-nucleotide phosphodiesterase activity (4 genes)
cytidine deaminase activity (8 genes)
cytidylate kinase activity (4 genes)
cytochrome-b5 reductase activity, acting on nad(p)h (6 genes)
cytochrome-c oxidase activity (25 genes)
diacylglycerol kinase activity (12 genes)
diacylglycerol o-acyltransferase activity (4 genes)
dodecenoyl-coa delta-isomerase activity (4 genes)
endopeptidase activity (107 genes)
atp-dependent peptidase activity (5 genes)
aminopeptidase activity (29 genes)
carboxypeptidase activity (33 genes)
metallocarboxypeptidase activity (25 genes)
serine-type carboxypeptidase activity (4 genes)
aspartic-type endopeptidase activity (26 genes)
cysteine-type endopeptidase activity (77 genes)
calcium-dependent cysteine-type endopeptidase activity (15 genes)
metalloendopeptidase activity (104 genes)
serine-type endopeptidase activity (181 genes)
threonine-type endopeptidase activity (21 genes)
enoyl-coa hydratase activity (8 genes)
epoxide hydrolase activity (4 genes)
estradiol 17-beta-dehydrogenase activity (13 genes)
ethanolamine kinase activity (4 genes)
exo-alpha-sialidase activity (4 genes)
fatty-acyl-coa synthase activity (6 genes)
ferroxidase activity (15 genes)
fructose-2,6-bisphosphate 2-phosphatase activity (5 genes)
fructose-bisphosphate aldolase activity (4 genes)
glucokinase activity (5 genes)
glutathione transferase activity (31 genes)
glycerol-3-phosphate o-acyltransferase activity (4 genes)
glycerol kinase activity (4 genes)
guanylate cyclase activity (11 genes)
guanylate kinase activity (4 genes)
helicase activity (123 genes)
hexokinase activity (5 genes)
histone acetyltransferase activity (47 genes)
histone deacetylase activity (26 genes)
hyalurononglucosaminidase activity (9 genes)
inorganic diphosphatase activity (4 genes)
1-phosphatidylinositol 4-kinase activity (4 genes)
phosphatidylinositol phospholipase c activity (17 genes)
phosphatidylinositol-3-phosphatase activity (10 genes)
phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity (8 genes)
inositol-polyphosphate 5-phosphatase activity (9 genes)
isocitrate dehydrogenase (nad+) activity (4 genes)
l-lactate dehydrogenase activity (4 genes)
leukotriene-c4 synthase activity (4 genes)
long-chain-acyl-coa dehydrogenase activity (4 genes)
long-chain fatty acid-coa ligase activity (13 genes)
lysine n-acetyltransferase activity, acting on acetyl phosphate as donor (6 genes)
methylenetetrahydrofolate dehydrogenase (nadp+) activity (4 genes)
monooxygenase activity (92 genes)
n,n-dimethylaniline monooxygenase activity (9 genes)
nuclease activity (136 genes)
endonuclease activity (85 genes)
endodeoxyribonuclease activity (9 genes)
endoribonuclease activity (34 genes)
rna-dna hybrid ribonuclease activity (4 genes)
ribonuclease iii activity (5 genes)
ribonuclease p activity (11 genes)
exonuclease activity (52 genes)
phosphodiesterase i activity (6 genes)
deoxyribonuclease i activity (5 genes)
exoribonuclease activity (4 genes)
poly(a)-specific ribonuclease activity (14 genes)
deoxyribonuclease activity (7 genes)
ribonuclease activity (32 genes)
nucleoside diphosphate kinase activity (18 genes)
nucleotide diphosphatase activity (6 genes)
hydrolase activity, hydrolyzing o-glycosyl compounds (26 genes)
alpha-amylase activity (5 genes)
alpha-1,4-glucosidase activity (4 genes)
alpha-mannosidase activity (7 genes)
beta-n-acetylhexosaminidase activity (5 genes)
beta-galactosidase activity (6 genes)
chitinase activity (8 genes)
mannosyl-oligosaccharide 1,2-alpha-mannosidase activity (7 genes)
dolichyl-diphosphooligosaccharide-protein glycotransferase activity (9 genes)
oxoglutarate dehydrogenase (succinyl-transferring) activity (4 genes)
pantothenate kinase activity (4 genes)
peptide alpha-n-acetyltransferase activity (11 genes)
peroxidase activity (37 genes)
glutathione peroxidase activity (22 genes)
phosphoglucomutase activity (6 genes)
phospholipase activity (18 genes)
lysophospholipase activity (17 genes)
phospholipase a2 activity (27 genes)
phospholipase c activity (8 genes)
phospholipase d activity (5 genes)
phosphopyruvate hydratase activity (5 genes)
phosphorylase activity (5 genes)
polynucleotide adenylyltransferase activity (9 genes)
polypeptide n-acetylgalactosaminyltransferase activity (17 genes)
procollagen-proline 4-dioxygenase activity (5 genes)
prenyltransferase activity (8 genes)
protein-arginine deiminase activity (5 genes)
protein kinase activity (537 genes)
protein serine/threonine kinase activity (430 genes)
transmembrane receptor protein serine/threonine kinase activity (13 genes)
amp-activated protein kinase activity (7 genes)
calmodulin-dependent protein kinase activity (23 genes)
myosin light chain kinase activity (4 genes)
phosphorylase kinase activity (5 genes)
camp-dependent protein kinase activity (4 genes)
cyclin-dependent protein serine/threonine kinase activity (33 genes)
eukaryotic translation initiation factor 2alpha kinase activity (4 genes)
protein kinase c activity (16 genes)
g-protein coupled receptor kinase activity (6 genes)
nf-kappab-inducing kinase activity (4 genes)
jun kinase kinase kinase activity (10 genes)
map kinase activity (14 genes)
map kinase kinase activity (12 genes)
map kinase kinase kinase activity (23 genes)
ribosomal protein s6 kinase activity (8 genes)
protein serine/threonine/tyrosine kinase activity (29 genes)
protein tyrosine kinase activity (115 genes)
transmembrane receptor protein tyrosine kinase activity (54 genes)
non-membrane spanning protein tyrosine kinase activity (45 genes)
protein-l-isoaspartate (d-aspartate) o-methyltransferase activity (4 genes)
protein-lysine 6-oxidase activity (5 genes)
phosphoprotein phosphatase activity (143 genes)
protein serine/threonine phosphatase activity (59 genes)
calcium-dependent protein serine/threonine phosphatase activity (5 genes)
protein tyrosine phosphatase activity (98 genes)
non-membrane spanning protein tyrosine phosphatase activity (6 genes)
pyruvate dehydrogenase (acetyl-transferring) kinase activity (4 genes)
retinol dehydrogenase activity (18 genes)
ribose phosphate diphosphokinase activity (6 genes)
serine c-palmitoyltransferase activity (5 genes)
sphingomyelin phosphodiesterase activity (8 genes)
stearoyl-coa 9-desaturase activity (6 genes)
steroid delta-isomerase activity (5 genes)
sterol esterase activity (14 genes)
superoxide dismutase activity (5 genes)
thioredoxin-disulfide reductase activity (7 genes)
thiosulfate sulfurtransferase activity (4 genes)
triglyceride lipase activity (27 genes)
aminoacyl-trna ligase activity (40 genes)
phenylalanine-trna ligase activity (4 genes)
ubiquitin-protein transferase activity (295 genes)
thiol-dependent ubiquitin-specific protease activity (89 genes)
uridine kinase activity (4 genes)
xanthine dehydrogenase activity (5 genes)
enzyme inhibitor activity (42 genes)
phospholipase inhibitor activity (5 genes)
protein kinase inhibitor activity (27 genes)
cyclin-dependent protein serine/threonine kinase inhibitor activity (12 genes)
camp-dependent protein kinase inhibitor activity (8 genes)
protein phosphatase inhibitor activity (34 genes)
protein serine/threonine phosphatase inhibitor activity (10 genes)
endopeptidase inhibitor activity (27 genes)
serine-type endopeptidase inhibitor activity (118 genes)
cysteine-type endopeptidase inhibitor activity (44 genes)
nuclear receptor activity (37 genes)
thyroid hormone receptor activity (5 genes)
transmembrane signaling receptor activity (219 genes)
gaba-a receptor activity (19 genes)
cytokine receptor activity (45 genes)
ciliary neurotrophic factor receptor activity (5 genes)
interleukin-1 receptor activity (6 genes)
g-protein coupled receptor activity (639 genes)
extracellularly atp-gated cation channel activity (7 genes)
adrenergic receptor activity (18 genes)
angiotensin type ii receptor activity (4 genes)
chemokine receptor activity (19 genes)
dopamine neurotransmitter receptor activity (5 genes)
prostaglandin receptor activity (5 genes)
prostaglandin e receptor activity (5 genes)
galanin receptor activity (10 genes)
histamine receptor activity (4 genes)
ionotropic glutamate receptor activity (18 genes)
ampa glutamate receptor activity (4 genes)
nmda glutamate receptor activity (7 genes)
leukotriene receptor activity (4 genes)
melanocortin receptor activity (5 genes)
n-formyl peptide receptor activity (7 genes)
neuropeptide y receptor activity (10 genes)
olfactory receptor activity (1111 genes)
opioid receptor activity (5 genes)
g-protein coupled serotonin receptor activity (39 genes)
somatostatin receptor activity (5 genes)
vasopressin receptor activity (7 genes)
transmembrane receptor protein tyrosine phosphatase activity (5 genes)
ephrin receptor activity (14 genes)
gpi-linked ephrin receptor activity (7 genes)
transmembrane-ephrin receptor activity (14 genes)
fibroblast growth factor-activated receptor activity (5 genes)
insulin-activated receptor activity (4 genes)
vascular endothelial growth factor-activated receptor activity (6 genes)
transforming growth factor beta-activated receptor activity (14 genes)
transforming growth factor beta receptor activity, type i (7 genes)
transforming growth factor beta receptor activity, type ii (5 genes)
neurotrophin receptor activity (4 genes)
tumor necrosis factor-activated receptor activity (6 genes)
low-density lipoprotein receptor activity (10 genes)
netrin receptor activity (7 genes)
scavenger receptor activity (44 genes)
nuclear export signal receptor activity (8 genes)
transmembrane receptor protein tyrosine kinase adaptor activity (9 genes)
sh3/sh2 adaptor activity (27 genes)
map-kinase scaffold activity (9 genes)
protein kinase c binding (62 genes)
guanyl-nucleotide exchange factor activity (160 genes)
arf guanyl-nucleotide exchange factor activity (16 genes)
ran guanyl-nucleotide exchange factor activity (4 genes)
ras guanyl-nucleotide exchange factor activity (13 genes)
rho guanyl-nucleotide exchange factor activity (67 genes)
gdp-dissociation inhibitor activity (13 genes)
gtpase inhibitor activity (4 genes)
gtpase activator activity (239 genes)
receptor binding (413 genes)
fibroblast growth factor receptor binding (25 genes)
type 1 fibroblast growth factor receptor binding (5 genes)
frizzled binding (43 genes)
type 2 fibroblast growth factor receptor binding (5 genes)
notch binding (23 genes)
patched binding (7 genes)
type ii transforming growth factor beta receptor binding (7 genes)
death receptor binding (15 genes)
scavenger receptor binding (4 genes)
cytokine activity (209 genes)
cytokine receptor binding (22 genes)
ciliary neurotrophic factor receptor binding (7 genes)
growth hormone receptor binding (5 genes)
type i interferon receptor binding (24 genes)
interleukin-2 receptor binding (5 genes)
interleukin-6 receptor binding (7 genes)
interleukin-12 receptor binding (6 genes)
prolactin receptor binding (27 genes)
interleukin-1 receptor binding (17 genes)
epidermal growth factor receptor binding (36 genes)
insulin receptor binding (26 genes)
insulin-like growth factor receptor binding (14 genes)
transforming growth factor beta receptor binding (44 genes)
platelet-derived growth factor receptor binding (14 genes)
tumor necrosis factor receptor binding (34 genes)
neurotrophin p75 receptor binding (6 genes)
neurotrophin trka receptor binding (6 genes)
vascular endothelial growth factor receptor binding (6 genes)
erbb-2 class receptor binding (4 genes)
integrin binding (120 genes)
hormone activity (118 genes)
neuropeptide hormone activity (25 genes)
pheromone activity (16 genes)
structural molecule activity (189 genes)
structural constituent of cytoskeleton (63 genes)
extracellular matrix structural constituent (113 genes)
structural constituent of eye lens (24 genes)
transporter activity (67 genes)
ion channel activity (183 genes)
intracellular cyclic nucleotide activated cation channel activity (5 genes)
intracellular camp activated cation channel activity (9 genes)
intracellular cgmp activated cation channel activity (7 genes)
volume-sensitive anion channel activity (6 genes)
calcium activated cation channel activity (14 genes)
intracellular calcium activated chloride channel activity (15 genes)
extracellular ligand-gated ion channel activity (43 genes)
extracellularly glutamate-gated ion channel activity (5 genes)
inhibitory extracellular ligand-gated ion channel activity (11 genes)
inward rectifier potassium channel activity (20 genes)
gap junction channel activity (13 genes)
voltage-gated ion channel activity (138 genes)
voltage-gated calcium channel activity (44 genes)
calcium channel regulator activity (34 genes)
voltage-gated chloride channel activity (11 genes)
voltage-gated sodium channel activity (19 genes)
voltage-gated potassium channel activity (64 genes)
delayed rectifier potassium channel activity (30 genes)
anion channel activity (4 genes)
chloride channel activity (57 genes)
cation channel activity (33 genes)
calcium channel activity (88 genes)
potassium channel activity (79 genes)
sodium channel activity (31 genes)
amine transmembrane transporter activity (4 genes)
l-glutamate transmembrane transporter activity (13 genes)
high-affinity glutamate transmembrane transporter activity (4 genes)
inorganic phosphate transmembrane transporter activity (5 genes)
lipid transporter activity (27 genes)
long-chain fatty acid transporter activity (13 genes)
neurotransmitter transporter activity (5 genes)
neurotransmitter:sodium symporter activity (19 genes)
dopamine:sodium symporter activity (4 genes)
gamma-aminobutyric acid:sodium symporter activity (4 genes)
nucleoside transmembrane transporter activity (8 genes)
oxygen carrier activity (12 genes)
atp transmembrane transporter activity (7 genes)
sugar:proton symporter activity (4 genes)
glucose transmembrane transporter activity (15 genes)
water transmembrane transporter activity (4 genes)
copper ion transmembrane transporter activity (6 genes)
iron ion transmembrane transporter activity (5 genes)
manganese ion transmembrane transporter activity (5 genes)
zinc ion transmembrane transporter activity (22 genes)
calcium-transporting atpase activity (11 genes)
sodium:potassium-exchanging atpase activity (11 genes)
glucose:sodium symporter activity (5 genes)
nucleoside:sodium symporter activity (4 genes)
calcium:sodium antiporter activity (5 genes)
inorganic anion exchanger activity (10 genes)
atp:adp antiporter activity (4 genes)
snap receptor activity (32 genes)
steroid binding (44 genes)
vitamin d binding (6 genes)
retinoid binding (8 genes)
fatty acid binding (29 genes)
iron ion binding (152 genes)
copper ion binding (58 genes)
calcium ion binding (613 genes)
protein binding (5084 genes)
calmodulin binding (184 genes)
collagen binding (65 genes)
insulin-like growth factor binding (23 genes)
lamin binding (16 genes)
profilin binding (12 genes)
tropomyosin binding (13 genes)
atp binding (1421 genes)
gtp binding (353 genes)
fk506 binding (4 genes)
galactose binding (6 genes)
glucose binding (12 genes)
mannose binding (26 genes)
glycosaminoglycan binding (25 genes)
hyaluronic acid binding (21 genes)
folic acid binding (13 genes)
phospholipid binding (104 genes)
calcium-dependent phospholipid binding (54 genes)
1-phosphatidylinositol binding (14 genes)
phosphatidylinositol-4,5-bisphosphate binding (76 genes)
phosphatidylinositol-3,4,5-trisphosphate binding (39 genes)
phospholipid transporter activity (22 genes)
odorant binding (362 genes)
pheromone binding (90 genes)
chemokine activity (43 genes)
beta-catenin binding (86 genes)
microtubule binding (228 genes)
g-protein coupled photoreceptor activity (9 genes)
protein c-terminus binding (239 genes)
atp-dependent helicase activity (12 genes)
monocarboxylic acid transmembrane transporter activity (13 genes)
high-density lipoprotein particle binding (9 genes)
axon guidance receptor activity (10 genes)
enzyme activator activity (31 genes)
calcium sensitive guanylate cyclase activator activity (4 genes)
chitin binding (11 genes)
glutamate receptor activity (15 genes)
n-acetyltransferase activity (18 genes)
phosphoric diester hydrolase activity (53 genes)
growth factor activity (147 genes)
cytoskeletal protein binding (98 genes)
cytoskeletal adaptor activity (7 genes)
dna-dependent atpase activity (35 genes)
5s rrna binding (13 genes)
alcohol dehydrogenase (nadp+) activity (18 genes)
ubiquinol-cytochrome-c reductase activity (7 genes)
primary amine oxidase activity (9 genes)
transcription factor binding (405 genes)
translation factor activity, rna binding (12 genes)
nadh dehydrogenase (ubiquinone) activity (29 genes)
protein tyrosine/serine/threonine phosphatase activity (40 genes)
nuclear localization sequence binding (21 genes)
camp response element binding protein binding (9 genes)
poly(a) binding (27 genes)
drug binding (109 genes)
sulfotransferase activity (48 genes)
protein phosphatase 1 binding (21 genes)
methyltransferase activity (168 genes)
o-methyltransferase activity (9 genes)
rna methyltransferase activity (9 genes)
trna methyltransferase activity (8 genes)
succinate dehydrogenase (ubiquinone) activity (5 genes)
adenylate cyclase binding (16 genes)
glycogen phosphorylase activity (4 genes)
rna-dependent atpase activity (4 genes)
neuropeptide receptor activity (12 genes)
eukaryotic initiation factor 4e binding (10 genes)
metalloendopeptidase inhibitor activity (13 genes)
udp-glycosyltransferase activity (21 genes)
phosphatidate phosphatase activity (13 genes)
ferrous iron binding (32 genes)
ferric iron binding (17 genes)
heparin binding (151 genes)
g-protein coupled amine receptor activity (15 genes)
peptidase activity (518 genes)
cysteine-type peptidase activity (139 genes)
metalloexopeptidase activity (11 genes)
serine-type peptidase activity (163 genes)
metallopeptidase activity (160 genes)
exopeptidase activity (7 genes)
dipeptidyl-peptidase activity (12 genes)
peptidyl-dipeptidase activity (4 genes)
trna-specific adenosine deaminase activity (7 genes)
5'-nucleotidase activity (11 genes)
poly(u) rna binding (29 genes)
zinc ion binding (685 genes)
secondary active sulfate transmembrane transporter activity (13 genes)
calcium, potassium:sodium antiporter activity (5 genes)
protein methyltransferase activity (13 genes)
lipid binding (289 genes)
calcium- and calmodulin-responsive adenylate cyclase activity (5 genes)
3'-5'-exodeoxyribonuclease activity (5 genes)
dna binding, bending (15 genes)
structural constituent of muscle (23 genes)
voltage-gated anion channel activity (5 genes)
single-stranded dna 3'-5' exodeoxyribonuclease activity (6 genes)
7s rna binding (6 genes)
protein transmembrane transporter activity (13 genes)
cation transmembrane transporter activity (18 genes)
methyl-cpg binding (21 genes)
signaling pattern recognition receptor activity (8 genes)
protein tyrosine/threonine phosphatase activity (4 genes)
high voltage-gated calcium channel activity (10 genes)
map kinase kinase kinase kinase activity (5 genes)
rna polymerase ii carboxy-terminal domain kinase activity (17 genes)
sialyltransferase activity (21 genes)
o-acyltransferase activity (10 genes)
acetylglucosaminyltransferase activity (24 genes)
acetylgalactosaminyltransferase activity (24 genes)
galactosyltransferase activity (21 genes)
thioredoxin peroxidase activity (6 genes)
mechanosensitive ion channel activity (11 genes)
testosterone 16-alpha-hydroxylase activity (5 genes)
arachidonic acid monooxygenase activity (6 genes)
arachidonic acid epoxygenase activity (34 genes)
steroid hydroxylase activity (52 genes)
3'-5' exonuclease activity (20 genes)
5'-3' exonuclease activity (12 genes)
fucosyltransferase activity (9 genes)
ctd phosphatase activity (6 genes)
beta-glucosidase activity (4 genes)
phosphatidylethanolamine binding (10 genes)
selenium binding (9 genes)
jun kinase binding (15 genes)
inositol-1,4,5-trisphosphate 3-kinase activity (4 genes)
[heparan sulfate]-glucosamine 3-sulfotransferase 1 activity (5 genes)
histone-arginine n-methyltransferase activity (9 genes)
palmitoyl-(protein) hydrolase activity (10 genes)
transaminase activity (22 genes)
sulfuric ester hydrolase activity (12 genes)
diphosphoinositol-polyphosphate diphosphatase activity (5 genes)
translation activator activity (10 genes)
udp-galactose:beta-n-acetylglucosamine beta-1,3-galactosyltransferase activity (7 genes)
melatonin receptor activity (4 genes)
benzodiazepine receptor activity (11 genes)
monoamine transmembrane transporter activity (8 genes)
bile acid:sodium symporter activity (5 genes)
anion transmembrane transporter activity (17 genes)
sodium:bicarbonate symporter activity (6 genes)
organic anion transmembrane transporter activity (19 genes)
folic acid transmembrane transporter activity (4 genes)
ammonium transmembrane transporter activity (7 genes)
phosphatidylcholine transporter activity (9 genes)
phosphatidylinositol transporter activity (9 genes)
taste receptor activity (16 genes)
g-protein coupled peptide receptor activity (35 genes)
n-acetyllactosaminide beta-1,3-n-acetylglucosaminyltransferase activity (9 genes)
ran gtpase binding (36 genes)
hydrogen-exporting atpase activity, phosphorylative mechanism (20 genes)
xenobiotic transmembrane transporting atpase activity (5 genes)
protein transporter activity (54 genes)
microtubule-severing atpase activity (9 genes)
atp-dependent microtubule motor activity, minus-end-directed (21 genes)
atp-dependent microtubule motor activity, plus-end-directed (28 genes)
camp-dependent protein kinase regulator activity (6 genes)
phosphorylase kinase regulator activity (4 genes)
ubiquitin-like modifier activating enzyme activity (10 genes)
rrna methyltransferase activity (6 genes)
cysteine-type endopeptidase activator activity involved in apoptotic process (10 genes)
n-acetylmuramoyl-l-alanine amidase activity (4 genes)
s-adenosylmethionine-dependent methyltransferase activity (15 genes)
crossover junction endodeoxyribonuclease activity (8 genes)
exodeoxyribonuclease iii activity (5 genes)
fructokinase activity (5 genes)
glycerophosphodiester phosphodiesterase activity (5 genes)
inositol monophosphate 1-phosphatase activity (4 genes)
oxaloacetate decarboxylase activity (4 genes)
phosphatidylcholine 1-acylhydrolase activity (6 genes)
trna (guanine-n1-)-methyltransferase activity (4 genes)
electron transfer activity (43 genes)
biotin binding (4 genes)
four-way junction helicase activity (5 genes)
photoreceptor activity (10 genes)
fatty acid elongase activity (7 genes)
calcium-dependent protein serine/threonine kinase activity (7 genes)
pseudouridine synthase activity (13 genes)
fmn binding (16 genes)
phosphatidylinositol-5-phosphate binding (15 genes)
double-stranded methylated dna binding (5 genes)
h4 histone acetyltransferase activity (12 genes)
telomerase inhibitor activity (6 genes)
anaphase-promoting complex binding (7 genes)
[heparan sulfate]-glucosamine n-sulfotransferase activity (4 genes)
glucuronosyltransferase activity (27 genes)
coreceptor activity (20 genes)
protein disulfide oxidoreductase activity (20 genes)
disulfide oxidoreductase activity (7 genes)
peptide disulfide oxidoreductase activity (8 genes)
thrombin-activated receptor activity (4 genes)
ion transmembrane transporter activity (8 genes)
hydrogen ion transmembrane transporter activity (30 genes)
sodium ion transmembrane transporter activity (5 genes)
ferrous iron transmembrane transporter activity (6 genes)
magnesium ion transmembrane transporter activity (16 genes)
organic cation transmembrane transporter activity (6 genes)
bicarbonate transmembrane transporter activity (14 genes)
chloride transmembrane transporter activity (16 genes)
phosphate ion transmembrane transporter activity (4 genes)
sulfate transmembrane transporter activity (15 genes)
bile acid transmembrane transporter activity (14 genes)
lactate transmembrane transporter activity (6 genes)
sialic acid transmembrane transporter activity (8 genes)
citrate transmembrane transporter activity (5 genes)
succinate transmembrane transporter activity (7 genes)
urate transmembrane transporter activity (4 genes)
pyrimidine nucleotide-sugar transmembrane transporter activity (7 genes)
amino acid transmembrane transporter activity (21 genes)
basic amino acid transmembrane transporter activity (4 genes)
neutral amino acid transmembrane transporter activity (12 genes)
l-amino acid transmembrane transporter activity (14 genes)
l-alanine transmembrane transporter activity (7 genes)
arginine transmembrane transporter activity (6 genes)
l-aspartate transmembrane transporter activity (6 genes)
l-glutamine transmembrane transporter activity (5 genes)
glycine transmembrane transporter activity (8 genes)
l-lysine transmembrane transporter activity (6 genes)
l-proline transmembrane transporter activity (8 genes)
choline transmembrane transporter activity (5 genes)
carnitine transmembrane transporter activity (4 genes)
acyl carnitine transmembrane transporter activity (5 genes)
heme transporter activity (5 genes)
drug transmembrane transporter activity (14 genes)
fatty acid transmembrane transporter activity (4 genes)
aminophospholipid transmembrane transporter activity (4 genes)
sterol transporter activity (13 genes)
water channel activity (12 genes)
proton channel activity (5 genes)
urea channel activity (5 genes)
channel activity (22 genes)
calcium-activated potassium channel activity (15 genes)
outward rectifier potassium channel activity (14 genes)
ligand-gated ion channel activity (11 genes)
kainate selective glutamate receptor activity (6 genes)
calcium-release channel activity (14 genes)
store-operated calcium channel activity (13 genes)
ligand-gated sodium channel activity (7 genes)
porin activity (6 genes)
symporter activity (108 genes)
antiporter activity (41 genes)
solute:proton antiporter activity (13 genes)
anion:anion antiporter activity (20 genes)
sodium-dependent phosphate transmembrane transporter activity (4 genes)
sodium-independent organic anion transmembrane transporter activity (17 genes)
thyroid hormone transmembrane transporter activity (5 genes)
cation:chloride symporter activity (6 genes)
potassium:chloride symporter activity (10 genes)
sodium:proton antiporter activity (11 genes)
potassium:proton antiporter activity (10 genes)
p-p-bond-hydrolysis-driven protein transmembrane transporter activity (5 genes)
potassium channel regulator activity (36 genes)
acetylcholine receptor activity (15 genes)
g-protein activated inward rectifier potassium channel activity (4 genes)
cholesterol binding (41 genes)
efflux transmembrane transporter activity (7 genes)
dna translocase activity (6 genes)
tubulin binding (58 genes)
toxic substance binding (13 genes)
fatty acid ligase activity (5 genes)
quaternary ammonium group transmembrane transporter activity (6 genes)
phospholipase activator activity (4 genes)
cyclosporin a binding (13 genes)
peptidoglycan receptor activity (4 genes)
nickel cation binding (5 genes)
nad(p)h oxidase activity (5 genes)
superoxide-generating nadph oxidase activity (8 genes)
superoxide-generating nadph oxidase activator activity (8 genes)
amp binding (18 genes)
antioxidant activity (25 genes)
kynurenine-oxoglutarate transaminase activity (4 genes)
steroid dehydrogenase activity (13 genes)
channel regulator activity (11 genes)
protein-arginine n-methyltransferase activity (8 genes)
protein-lysine n-methyltransferase activity (20 genes)
small conductance calcium-activated potassium channel activity (5 genes)
palmitoyl-coa hydrolase activity (17 genes)
lipase activity (21 genes)
kinase activity (623 genes)
1-phosphatidylinositol-3-kinase activity (10 genes)
phosphatidylinositol phosphate kinase activity (8 genes)
1-phosphatidylinositol-4-phosphate 5-kinase activity (7 genes)
activin receptor activity, type i (4 genes)
acetyltransferase activity (19 genes)
palmitoyltransferase activity (19 genes)
n-acyltransferase activity (5 genes)
trna (cytosine-5-)-methyltransferase activity (6 genes)
pyrophosphatase activity (7 genes)
oxidoreductase activity (599 genes)
c-c chemokine receptor activity (24 genes)
c-x-c chemokine receptor activity (8 genes)
protein-hormone receptor activity (9 genes)
pheromone receptor activity (99 genes)
peptidase activator activity (11 genes)
superoxide dismutase copper chaperone activity (5 genes)
cyclin-dependent protein serine/threonine kinase regulator activity (31 genes)
glycine binding (15 genes)
glutamate binding (14 genes)
amino acid binding (32 genes)
oxidoreductase activity, acting on the ch-oh group of donors, nad or nadp as acceptor (32 genes)
oxidoreductase activity, acting on the aldehyde or oxo group of donors, nad or nadp as acceptor (24 genes)
oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor (5 genes)
oxidoreductase activity, acting on the ch-ch group of donors (25 genes)
oxidoreductase activity, acting on the ch-ch group of donors, nad or nadp as acceptor (5 genes)
oxidoreductase activity, acting on the ch-nh2 group of donors, oxygen as acceptor (5 genes)
oxidoreductase activity, acting on nad(p)h (13 genes)
oxidoreductase activity, acting on nad(p)h, quinone or similar compound as acceptor (6 genes)
oxidoreductase activity, acting on a sulfur group of donors, nad(p) as acceptor (6 genes)
oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor (6 genes)
oxidoreductase activity, acting on peroxide as acceptor (5 genes)
oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen (19 genes)
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen (78 genes)
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors (16 genes)
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, nad(p)h as one donor, and incorporation of one atom of oxygen (7 genes)
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen (58 genes)
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen (4 genes)
oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen (4 genes)
oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water (5 genes)
transferase activity (1722 genes)
hydroxymethyl-, formyl- and related transferase activity (4 genes)
transferase activity, transferring acyl groups (166 genes)
transferase activity, transferring acyl groups other than amino-acyl groups (18 genes)
transferase activity, transferring glycosyl groups (204 genes)
transferase activity, transferring hexosyl groups (23 genes)
transferase activity, transferring pentosyl groups (8 genes)
transferase activity, transferring alkyl or aryl (other than methyl) groups (7 genes)
transferase activity, transferring phosphorus-containing groups (11 genes)
phosphotransferase activity, alcohol group as acceptor (20 genes)
phosphotransferase activity, phosphate group as acceptor (4 genes)
nucleotidyltransferase activity (78 genes)
phosphotransferase activity, for other substituted phosphate groups (8 genes)
sulfurtransferase activity (4 genes)
hydrolase activity (1574 genes)
hydrolase activity, acting on ester bonds (30 genes)
thiolester hydrolase activity (14 genes)
phosphatase activity (128 genes)
hydrolase activity, acting on glycosyl bonds (76 genes)
hydrolase activity, hydrolyzing n-glycosyl compounds (6 genes)
dipeptidase activity (14 genes)
cysteine-type carboxypeptidase activity (4 genes)
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds (13 genes)
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides (25 genes)
hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines (6 genes)
hydrolase activity, acting on acid anhydrides (8 genes)
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides (10 genes)
lyase activity (145 genes)
carboxy-lyase activity (24 genes)
phosphorus-oxygen lyase activity (20 genes)
isomerase activity (114 genes)
racemase and epimerase activity, acting on carbohydrates and derivatives (5 genes)
intramolecular transferase activity, phosphotransferases (8 genes)
ligase activity (133 genes)
atpase activity (225 genes)
g-protein coupled acetylcholine receptor activity (7 genes)
gaba receptor activity (4 genes)
retinal binding (12 genes)
ligand-dependent nuclear receptor binding (24 genes)
extracellularly glycine-gated chloride channel activity (6 genes)
galactoside binding (6 genes)
thiol oxidase activity (4 genes)
ras gtpase binding (22 genes)
map kinase tyrosine/serine/threonine phosphatase activity (13 genes)
myosin binding (28 genes)
tbp-class protein binding (22 genes)
rap guanyl-nucleotide exchange factor activity (5 genes)
ceramidase activity (4 genes)
peptide hormone binding (44 genes)
rho gtpase binding (46 genes)
gtp-rho binding (18 genes)
structural constituent of nuclear pore (23 genes)
snrna binding (11 genes)
u6 snrna binding (9 genes)
syntaxin-1 binding (24 genes)
purine nucleotide binding (5 genes)
sodium channel regulator activity (28 genes)
chloride channel regulator activity (4 genes)
au-rich element binding (19 genes)
5'-flap endonuclease activity (7 genes)
nucleoside-diphosphatase activity (13 genes)
nucleoside-triphosphatase activity (11 genes)
rab guanyl-nucleotide exchange factor activity (36 genes)
sh3 domain binding (131 genes)
cholesterol transporter activity (19 genes)
phospholipid scramblase activity (11 genes)
fibroblast growth factor binding (26 genes)
rab gtpase binding (142 genes)
wnt-protein binding (37 genes)
trna dihydrouridine synthase activity (4 genes)
dead/h-box rna helicase binding (8 genes)
sodium:dicarboxylate symporter activity (4 genes)
semaphorin receptor activity (11 genes)
ral gtpase binding (13 genes)
aryl hydrocarbon receptor binding (11 genes)
vinculin binding (12 genes)
serine hydrolase activity (6 genes)
phosphatidylinositol n-acetylglucosaminyltransferase activity (6 genes)
histone-lysine n-methyltransferase activity (32 genes)
alkane 1-monooxygenase activity (6 genes)
3-hydroxyacyl-coa dehydratase activity (5 genes)
guanyl nucleotide binding (18 genes)
gdp binding (64 genes)
pyrimidine nucleotide binding (4 genes)
dna n-glycosylase activity (8 genes)
mannokinase activity (5 genes)
nucleobase-containing compound kinase activity (8 genes)
nucleoside kinase activity (10 genes)
kinase regulator activity (4 genes)
phosphatase regulator activity (8 genes)
kinase activator activity (7 genes)
kinase inhibitor activity (5 genes)
phosphatase inhibitor activity (5 genes)
deacetylase activity (15 genes)
intermediate filament binding (10 genes)
centromeric dna binding (4 genes)
deaminase activity (6 genes)
oxalate transmembrane transporter activity (11 genes)
toxin transmembrane transporter activity (6 genes)
protein-cysteine s-palmitoyltransferase activity (28 genes)
ige receptor activity (4 genes)
nedd8-specific protease activity (6 genes)
ubiquitin-like protein transferase activity (4 genes)
nedd8 transferase activity (5 genes)
sumo transferase activity (21 genes)
oxygen binding (23 genes)
cation-transporting atpase activity (4 genes)
phospholipase a2 inhibitor activity (4 genes)
growth factor binding (47 genes)
isoprenoid binding (6 genes)
retinol binding (13 genes)
vitamin binding (5 genes)
rrna binding (45 genes)
calcium channel inhibitor activity (9 genes)
ige binding (6 genes)
igg binding (8 genes)
immunoglobulin binding (4 genes)
chloride channel inhibitor activity (11 genes)
potassium channel inhibitor activity (9 genes)
sodium channel inhibitor activity (8 genes)
protein kinase regulator activity (9 genes)
protein phosphatase regulator activity (32 genes)
kinesin binding (49 genes)
enzyme binding (426 genes)
kinase binding (106 genes)
protein kinase binding (527 genes)
phosphatase binding (52 genes)
protein phosphatase binding (108 genes)
protein domain specific binding (338 genes)
syntaxin binding (66 genes)
structural constituent of myelin sheath (10 genes)
cytokine binding (41 genes)
chemokine binding (27 genes)
c-c chemokine binding (25 genes)
c-x3-c chemokine binding (5 genes)
interleukin-1 binding (5 genes)
interleukin-2 binding (4 genes)
diacylglycerol binding (8 genes)
heme binding (153 genes)
transmitter-gated ion channel activity (4 genes)
potassium ion leak channel activity (15 genes)
voltage-gated cation channel activity (6 genes)
acetylcholine-gated cation-selective channel activity (16 genes)
glutamate-gated calcium ion channel activity (5 genes)
gaba-gated chloride ion channel activity (12 genes)
transmembrane transporter activity (108 genes)
mhc class ii protein complex binding (6 genes)
extracellular matrix structural constituent conferring compression resistance (12 genes)
extracellular matrix constituent conferring elasticity (6 genes)
manganese ion binding (51 genes)
molybdenum ion binding (7 genes)
receptor signaling complex scaffold activity (14 genes)
gkap/homer scaffold activity (4 genes)
pdz domain binding (119 genes)
low-density lipoprotein particle binding (16 genes)
pyridoxal phosphate binding (53 genes)
semaphorin receptor binding (22 genes)
very-low-density lipoprotein particle receptor activity (4 genes)
enzyme regulator activity (24 genes)
nitric-oxide synthase regulator activity (7 genes)
carbohydrate binding (201 genes)
polysaccharide binding (15 genes)
guanylate cyclase regulator activity (4 genes)
lim domain binding (7 genes)
lrr domain binding (6 genes)
clathrin binding (40 genes)
structural constituent of epidermis (8 genes)
estrogen receptor activity (5 genes)
protein serine/threonine kinase inhibitor activity (8 genes)
protein kinase activator activity (27 genes)
protein tyrosine kinase activator activity (9 genes)
transmembrane receptor protein tyrosine kinase activator activity (4 genes)
adp-ribosylation factor binding (8 genes)
estrogen receptor binding (49 genes)
cyclin binding (32 genes)
protein phosphatase 2b binding (10 genes)
interleukin-17 receptor activity (5 genes)
translation repressor activity (12 genes)
ligand-dependent nuclear receptor transcription coactivator activity (60 genes)
thyroid hormone receptor coactivator activity (6 genes)
peptidase inhibitor activity (121 genes)
hemoglobin binding (5 genes)
ankyrin binding (23 genes)
spectrin binding (32 genes)
snorna binding (22 genes)
hsp70 protein binding (44 genes)
acetylcholine receptor regulator activity (7 genes)
acetylcholine receptor inhibitor activity (7 genes)
camp binding (22 genes)
cgmp binding (16 genes)
neurotransmitter receptor activity (40 genes)
u1 snrna binding (6 genes)
u2 snrna binding (6 genes)
u4 snrna binding (5 genes)
pre-mrna 3'-splice site binding (7 genes)
protein binding, bridging (72 genes)
rac guanyl-nucleotide exchange factor activity (17 genes)
gtpase regulator activity (15 genes)
gtp-dependent protein binding (25 genes)
beta-2-microglobulin binding (11 genes)
actin-dependent atpase activity (11 genes)
calcium-dependent atpase activity (5 genes)
tpr domain binding (8 genes)
endoplasmic reticulum signal peptide binding (4 genes)
mitochondrion targeting sequence binding (4 genes)
potassium ion binding (11 genes)
tat protein binding (9 genes)
receptor tyrosine kinase binding (64 genes)
thiamine pyrophosphate binding (5 genes)
mismatched dna binding (10 genes)
kininogen binding (4 genes)
filamin binding (17 genes)
troponin i binding (5 genes)
troponin t binding (4 genes)
heat shock protein binding (76 genes)
phosphatidylcholine binding (24 genes)
small gtpase binding (19 genes)
translation initiation factor binding (26 genes)
protein tag (13 genes)
sodium ion binding (13 genes)
chloride ion binding (11 genes)
carboxylic acid binding (12 genes)
l-ascorbic acid binding (19 genes)
cobalamin binding (9 genes)
titin binding (9 genes)
telethonin binding (6 genes)
mitogen-activated protein kinase kinase binding (19 genes)
mitogen-activated protein kinase kinase kinase binding (27 genes)
myosin v binding (21 genes)
chromatin dna binding (77 genes)
nucleosome binding (25 genes)
nucleosomal dna binding (23 genes)
nucleosomal histone binding (4 genes)
thyrotropin-releasing hormone receptor binding (4 genes)
peptidyl-proline 4-dioxygenase activity (7 genes)
polyubiquitin modification-dependent protein binding (28 genes)
ubiquitin conjugating enzyme binding (33 genes)
ubiquitin protein ligase binding (305 genes)
g-protein beta-subunit binding (13 genes)
g-protein beta/gamma-subunit complex binding (24 genes)
alpha-2a adrenergic receptor binding (4 genes)
beta-1 adrenergic receptor binding (4 genes)
beta-2 adrenergic receptor binding (8 genes)
type 1 angiotensin receptor binding (7 genes)
haptoglobin binding (11 genes)
hemoglobin alpha binding (6 genes)
ccr2 chemokine receptor binding (4 genes)
ccr3 chemokine receptor binding (4 genes)
ccr5 chemokine receptor binding (7 genes)
ccr6 chemokine receptor binding (12 genes)
d1 dopamine receptor binding (13 genes)
d2 dopamine receptor binding (6 genes)
d3 dopamine receptor binding (4 genes)
type 3 melanocortin receptor binding (5 genes)
type 4 melanocortin receptor binding (5 genes)
type 5 metabotropic glutamate receptor binding (6 genes)
g-protein coupled serotonin receptor binding (4 genes)
olfactory receptor binding (5 genes)
mu-type opioid receptor binding (4 genes)
very long-chain fatty acid-coa ligase activity (9 genes)
insulin-like growth factor i binding (12 genes)
insulin-like growth factor ii binding (8 genes)
thioesterase binding (11 genes)
myosin light chain binding (5 genes)
myosin heavy chain binding (8 genes)
nad-dependent histone deacetylase activity (h3-k14 specific) (12 genes)
clathrin heavy chain binding (7 genes)
clathrin light chain binding (6 genes)
bile acid binding (6 genes)
guanine/thymine mispair binding (6 genes)
single thymine insertion binding (5 genes)
ubiquitin-like protein binding (5 genes)
sumo binding (10 genes)
sumo polymer binding (5 genes)
phosphatidylinositol-3-phosphate binding (39 genes)
oxidized purine dna binding (7 genes)
mutlalpha complex binding (6 genes)
mutsalpha complex binding (8 genes)
purine-rich negative regulatory element binding (4 genes)
demethylase activity (9 genes)
histone demethylase activity (11 genes)
histone demethylase activity (h3-k4 specific) (5 genes)
histone demethylase activity (h3-k9 specific) (13 genes)
adenyl ribonucleotide binding (5 genes)
datp binding (4 genes)
gtpase activating protein binding (18 genes)
heterotrimeric g-protein binding (7 genes)
tumor necrosis factor receptor superfamily binding (6 genes)
palmitoyl-coa 9-desaturase activity (4 genes)
sterol binding (12 genes)
protein complex scaffold activity (68 genes)
bitter taste receptor activity (29 genes)
acetylcholine receptor binding (15 genes)
progesterone receptor binding (4 genes)
sphingomyelin synthase activity (4 genes)
calmodulin-dependent protein phosphatase activity (5 genes)
choline binding (6 genes)
dehydroascorbic acid transmembrane transporter activity (4 genes)
protein deacetylase activity (11 genes)
rna strand annealing activity (5 genes)
receptor serine/threonine kinase binding (8 genes)
activating transcription factor binding (39 genes)
sialic acid binding (9 genes)
peptide-methionine (r)-s-oxide reductase activity (4 genes)
poly(g) binding (10 genes)
estrogen response element binding (4 genes)
dna polymerase activity (4 genes)
apolipoprotein binding (15 genes)
apolipoprotein a-i binding (4 genes)
apolipoprotein receptor binding (4 genes)
gpi anchor binding (4 genes)
protein kinase a catalytic subunit binding (15 genes)
protein kinase a regulatory subunit binding (21 genes)
bis(5'-adenosyl)-hexaphosphatase activity (5 genes)
bis(5'-adenosyl)-pentaphosphatase activity (5 genes)
ubiquitin-ubiquitin ligase activity (11 genes)
dynactin binding (10 genes)
atp-dependent 3'-5' rna helicase activity (18 genes)
u3 snorna binding (6 genes)
box h/aca snorna binding (4 genes)
pirna binding (6 genes)
phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity (4 genes)
pyruvate dehydrogenase (nad+) activity (6 genes)
arginine binding (6 genes)
histone demethylase activity (h3-trimethyl-k4 specific) (4 genes)
histone demethylase activity (h3-dimethyl-k4 specific) (4 genes)
inhibin binding (7 genes)
type i transforming growth factor beta receptor binding (12 genes)
type iii transforming growth factor beta receptor binding (4 genes)
caffeine oxidase activity (6 genes)
immunoglobulin receptor binding (8 genes)
phosphatidylinositol 3-kinase activity (6 genes)
1-phosphatidylinositol-4-phosphate 3-kinase activity (7 genes)
phosphatidylinositol 3-kinase regulator activity (8 genes)
histone acetyltransferase binding (34 genes)
methylated histone binding (90 genes)
phosphatidylinositol binding (99 genes)
histone kinase activity (4 genes)
histone serine kinase activity (5 genes)
sirna binding (10 genes)
mirna binding (29 genes)
dopamine binding (16 genes)
protein-arginine omega-n monomethyltransferase activity (5 genes)
protein-arginine omega-n asymmetric methyltransferase activity (8 genes)
protein-arginine omega-n symmetric methyltransferase activity (4 genes)
udp-xylosyltransferase activity (4 genes)
glutamate receptor binding (15 genes)
ionotropic glutamate receptor binding (44 genes)
nuclear hormone receptor binding (26 genes)
glucocorticoid receptor binding (20 genes)
ethanol binding (4 genes)
5'-3' exodeoxyribonuclease activity (4 genes)
toll-like receptor binding (7 genes)
enhancer binding (12 genes)
selenocysteine insertion sequence binding (6 genes)
chondroitin sulfate binding (9 genes)
lipase binding (6 genes)
camp response element binding (13 genes)
nadh pyrophosphatase activity (7 genes)
signaling adaptor activity (13 genes)
ap-2 adaptor complex binding (10 genes)
rna stem-loop binding (15 genes)
snrna stem-loop binding (4 genes)
clathrin adaptor activity (9 genes)
ap-1 adaptor complex binding (5 genes)
toll-like receptor 4 binding (5 genes)
oligopeptide transmembrane transporter activity (7 genes)
lysophosphatidic acid binding (5 genes)
s-nitrosoglutathione binding (4 genes)
dinitrosyl-iron complex binding (4 genes)
mrna 3'-utr au-rich region binding (16 genes)
microsatellite binding (4 genes)
pre-mrna binding (14 genes)
gaf domain binding (4 genes)
mediator complex binding (5 genes)
long-chain fatty acid binding (5 genes)
long-chain fatty acyl-coa binding (7 genes)
small molecule binding (20 genes)
bmp binding (12 genes)
annealing helicase activity (7 genes)
phosphatidylinositol 3-kinase regulatory subunit binding (15 genes)
glutathione hydrolase activity (4 genes)
proteasome-activating atpase activity (6 genes)
k48-linked polyubiquitin modification-dependent protein binding (8 genes)
thiol-dependent ubiquitinyl hydrolase activity (86 genes)
signaling receptor activity (183 genes)
cargo receptor activity (10 genes)
sphingosine-1-phosphate receptor activity (8 genes)
collagen receptor activity (4 genes)
vascular endothelial growth factor binding (5 genes)
nodal binding (4 genes)
neuregulin binding (6 genes)
neuropilin binding (20 genes)
co-receptor binding (12 genes)
atpase inhibitor activity (6 genes)
neurexin family protein binding (13 genes)
histone methyltransferase activity (13 genes)
chemoattractant activity (37 genes)
rrna primary transcript binding (5 genes)
telomeric dna binding (28 genes)
neurotransmitter binding (11 genes)
acetylcholine binding (16 genes)
sh2 domain binding (37 genes)
lysophosphatidic acid acyltransferase activity (5 genes)
peptide binding (75 genes)
mhc class i protein binding (19 genes)
mhc class ii protein binding (7 genes)
phosphate ion binding (11 genes)
chemokine receptor binding (11 genes)
histone binding (123 genes)
aspartic endopeptidase activity, intramembrane cleaving (7 genes)
hormone binding (24 genes)
lipid phosphatase activity (10 genes)
phosphoric ester hydrolase activity (7 genes)
peptide antigen binding (36 genes)
t cell receptor binding (16 genes)
cd4 receptor binding (8 genes)
atpase activity, coupled (17 genes)
atpase activity, coupled to transmembrane movement of substances (55 genes)
ph domain binding (6 genes)
histone methyltransferase activity (h3-k4 specific) (14 genes)
identical protein binding (1203 genes)
protein homodimerization activity (920 genes)
actinin binding (13 genes)
vitamin d receptor binding (8 genes)
wnt-activated receptor activity (16 genes)
histone deacetylase binding (124 genes)
peptidoglycan binding (16 genes)
neuropeptide binding (23 genes)
retinoic acid receptor binding (20 genes)
peroxisome proliferator activated receptor binding (20 genes)
atp-dependent protein binding (11 genes)
alpha-tubulin binding (35 genes)
gamma-tubulin binding (30 genes)
ribonucleoprotein complex binding (45 genes)
ribosome binding (59 genes)
ribosomal large subunit binding (12 genes)
ribosomal small subunit binding (14 genes)
cysteine-type endopeptidase inhibitor activity involved in apoptotic process (28 genes)
single-stranded telomeric dna binding (7 genes)
neurotrophin binding (4 genes)
ubiquitin binding (68 genes)
3'-5' dna helicase activity (7 genes)
atp-dependent 3'-5' dna helicase activity (10 genes)
atp-dependent 5'-3' dna helicase activity (6 genes)
single-stranded dna-dependent atpase activity (12 genes)
ion binding (4 genes)
cation binding (13 genes)
organic acid binding (11 genes)
vascular endothelial growth factor receptor 2 binding (6 genes)
vascular endothelial growth factor receptor 3 binding (5 genes)
sulfate binding (5 genes)
glycosphingolipid binding (6 genes)
laminin binding (27 genes)
laminin-1 binding (6 genes)
adenosine-diphosphatase activity (5 genes)
phospholipase binding (20 genes)
glutathione binding (19 genes)
phosphatidylinositol-3,4-bisphosphate binding (25 genes)
proteoglycan binding (12 genes)
heparan sulfate proteoglycan binding (17 genes)
protein kinase b binding (18 genes)
bhlh transcription factor binding (27 genes)
mrf binding (6 genes)
protein membrane anchor (13 genes)
kinetochore binding (6 genes)
leucine zipper domain binding (11 genes)
adp binding (40 genes)
angiostatin binding (5 genes)
inositol 1,3,4,5 tetrakisphosphate binding (4 genes)
protein serine/threonine kinase activator activity (23 genes)
molybdopterin cofactor binding (6 genes)
phosphatidylinositol 3-kinase binding (28 genes)
insulin binding (7 genes)
insulin receptor substrate binding (11 genes)
sequence-specific dna binding (638 genes)
protein self-association (65 genes)
phosphatidylinositol-4-phosphate phosphatase activity (5 genes)
histone acetyltransferase activity (h4-k5 specific) (10 genes)
histone acetyltransferase activity (h4-k8 specific) (10 genes)
histone methyltransferase activity (h4-r3 specific) (4 genes)
protein binding involved in protein folding (22 genes)
transcription regulatory region dna binding (253 genes)
retinoic acid-responsive element binding (5 genes)
ion channel binding (138 genes)
small protein activating enzyme binding (5 genes)
ubiquitin-like protein ligase binding (20 genes)
ubiquitin-like protein conjugating enzyme binding (6 genes)
dna topoisomerase binding (5 genes)
s100 protein binding (13 genes)
macromolecular complex binding (435 genes)
g-protein coupled purinergic nucleotide receptor activity (12 genes)
uridine-diphosphatase activity (5 genes)
single-stranded dna 5'-3' exodeoxyribonuclease activity (4 genes)
myosin ii binding (5 genes)
translation regulator activity (22 genes)
alpha-catenin binding (12 genes)
gamma-catenin binding (14 genes)
cadherin binding (58 genes)
protein phosphorylated amino acid binding (4 genes)
unmethylated cpg binding (5 genes)
mercury ion binding (4 genes)
chemorepellent activity (28 genes)
dynein light chain binding (30 genes)
dynein heavy chain binding (15 genes)
dynein intermediate chain binding (33 genes)
syndecan binding (8 genes)
smad binding (62 genes)
glucosyltransferase activity (5 genes)
sphingolipid binding (5 genes)
natural killer cell lectin-like receptor binding (18 genes)
virion binding (7 genes)
hydroxyapatite binding (6 genes)
metal ion binding (3088 genes)
metal ion transmembrane transporter activity (15 genes)
ephrin receptor binding (31 genes)
transition metal ion binding (15 genes)
alpha-(1->3)-fucosyltransferase activity (5 genes)
er retention sequence binding (4 genes)
proton-transporting atp synthase activity, rotational mechanism (14 genes)
phosphatidylinositol-4,5-bisphosphate 3-kinase activity (5 genes)
1-phosphatidylinositol-3-kinase regulator activity (15 genes)
proton-transporting atpase activity, rotational mechanism (22 genes)
retinoid x receptor binding (22 genes)
thyroid hormone receptor binding (25 genes)
histone acetyltransferase activity (h4-k16 specific) (10 genes)
histone methyltransferase activity (h3-k9 specific) (6 genes)
histone methyltransferase activity (h3-k36 specific) (5 genes)
histone methyltransferase activity (h3-k27 specific) (7 genes)
tap binding (8 genes)
protein heterodimerization activity (575 genes)
protein dimerization activity (202 genes)
androsterone dehydrogenase activity (5 genes)
testosterone dehydrogenase (nad+) activity (6 genes)
ketosteroid monooxygenase activity (10 genes)
1-acylglycerophosphocholine o-acyltransferase activity (6 genes)
glucuronosyl-n-acetylgalactosaminyl-proteoglycan 4-beta-n-acetylgalactosaminyltransferase activity (4 genes)
acylglycerol lipase activity (11 genes)
alkylglycerophosphoethanolamine phosphodiesterase activity (6 genes)
nucleoside-triphosphate diphosphatase activity (6 genes)
protein n-terminus binding (130 genes)
ceramide cholinephosphotransferase activity (4 genes)
calcium-dependent phospholipase a2 activity (15 genes)
calcium-independent phospholipase a2 activity (6 genes)
3',5'-cyclic-gmp phosphodiesterase activity (18 genes)
acyl-coa hydrolase activity (17 genes)
adp-ribose diphosphatase activity (6 genes)
beta-adrenergic receptor kinase activity (4 genes)
bile-salt sulfotransferase activity (6 genes)
butyrate-coa ligase activity (6 genes)
dolichyldiphosphatase activity (5 genes)
receptor ligand activity (29 genes)
receptor antagonist activity (11 genes)
ccr chemokine receptor binding (25 genes)
mrna 5'-utr binding (23 genes)
monosaccharide binding (21 genes)
cofactor binding (21 genes)
quinone binding (10 genes)
ubiquinone binding (7 genes)
tau protein binding (21 genes)
activin binding (12 genes)
mitogen-activated protein kinase p38 binding (7 genes)
calcium-dependent protein binding (70 genes)
rac gtpase binding (55 genes)
brain-derived neurotrophic factor binding (5 genes)
nerve growth factor binding (8 genes)
platelet-derived growth factor binding (12 genes)
beta-tubulin binding (42 genes)
roundabout binding (5 genes)
calcium-induced calcium release activity (4 genes)
leukotriene-b4 20-monooxygenase activity (4 genes)
m7g(5')pppn diphosphatase activity (8 genes)
nucleoside phosphate kinase activity (5 genes)
sphingosine n-acyltransferase activity (7 genes)
steroid sulfotransferase activity (7 genes)
tau-protein kinase activity (20 genes)
transforming growth factor beta binding (21 genes)
hyaluronan synthase activity (4 genes)
arachidonic acid binding (9 genes)
testosterone 6-beta-hydroxylase activity (8 genes)
3'-phosphoadenosine 5'-phosphosulfate binding (7 genes)
flavin adenine dinucleotide binding (69 genes)
nadp binding (34 genes)
coenzyme binding (10 genes)
androgen receptor binding (30 genes)
dbd domain binding (5 genes)
lbd domain binding (7 genes)
ww domain binding (31 genes)
card domain binding (11 genes)
rs domain binding (7 genes)
low-density lipoprotein particle receptor binding (26 genes)
rage receptor binding (8 genes)
gaba receptor binding (15 genes)
phosphoserine residue binding (4 genes)
pyruvate transmembrane transporter activity (4 genes)
cell adhesion molecule binding (69 genes)
extracellular matrix binding (30 genes)
cobalt ion binding (10 genes)
nitric-oxide synthase binding (22 genes)
microtubule plus-end binding (16 genes)
microtubule minus-end binding (10 genes)
actin filament binding (193 genes)
protein kinase a binding (27 genes)
mitogen-activated protein kinase binding (29 genes)
gtpase binding (42 genes)
gdp-dissociation inhibitor binding (4 genes)
rho gdp-dissociation inhibitor binding (5 genes)
nf-kappab binding (32 genes)
unfolded protein binding (94 genes)
chaperone binding (98 genes)
atpase binding (94 genes)
dioxygenase activity (84 genes)
phosphoprotein binding (59 genes)
nad binding (47 genes)
muscle alpha-actinin binding (15 genes)
fatz binding (5 genes)
serotonin binding (12 genes)
epinephrine binding (8 genes)
norepinephrine binding (5 genes)
alpha-actinin binding (21 genes)
bh domain binding (11 genes)
ptb domain binding (4 genes)
hormone receptor binding (5 genes)
peptide hormone receptor binding (10 genes)
corticotropin-releasing hormone receptor 1 binding (4 genes)
corticotropin-releasing hormone receptor 2 binding (4 genes)
bh3 domain binding (5 genes)
nfat protein binding (5 genes)
iron-sulfur cluster binding (62 genes)
2 iron, 2 sulfur cluster binding (25 genes)
4 iron, 4 sulfur cluster binding (39 genes)
5'-deoxyribose-5-phosphate lyase activity (6 genes)
protein phosphatase 2a binding (36 genes)
misfolded protein binding (21 genes)
glycolipid binding (5 genes)
histone demethylase activity (h3-k36 specific) (10 genes)
hsp90 protein binding (42 genes)
g-quadruplex dna binding (8 genes)
peroxiredoxin activity (9 genes)
dynein light intermediate chain binding (33 genes)
diamine oxidase activity (4 genes)
phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity (4 genes)
nadp-retinol dehydrogenase activity (4 genes)
inositol-1,4,5-trisphosphate 5-phosphatase activity (4 genes)
carboxylic ester hydrolase activity (62 genes)
inositol hexakisphosphate 1-kinase activity (6 genes)
inositol hexakisphosphate 3-kinase activity (6 genes)
exo-alpha-(2->3)-sialidase activity (4 genes)
exo-alpha-(2->6)-sialidase activity (4 genes)
exo-alpha-(2->8)-sialidase activity (4 genes)
inositol monophosphate 3-phosphatase activity (4 genes)
inositol monophosphate 4-phosphatase activity (4 genes)
inositol monophosphate phosphatase activity (4 genes)
phosphatidylinositol phosphate phosphatase activity (4 genes)
d-glucose transmembrane transporter activity (4 genes)
lipase inhibitor activity (5 genes)
c3hc4-type ring finger domain binding (5 genes)
molecular adaptor activity (6 genes)
phosphatidylcholine-sterol o-acyltransferase activator activity (5 genes)
endopeptidase activator activity (7 genes)
cyclin-dependent protein serine/threonine kinase activator activity (8 genes)
lys63-specific deubiquitinase activity (11 genes)
rna polymerase ii sequence-specific dna binding transcription factor binding (7 genes)
ubiquitin protein ligase activity (218 genes)
ubiquitin conjugating enzyme activity (39 genes)
sumo ligase activity (4 genes)
importin-alpha family protein binding (7 genes)
telomeric d-loop binding (4 genes)
metalloaminopeptidase activity (16 genes)
oligopeptidase activity (4 genes)
armadillo repeat domain binding (10 genes)
telomerase rna binding (17 genes)
fibrinogen binding (5 genes)
collagen v binding (4 genes)
thrombospondin receptor activity (4 genes)
fructose binding (5 genes)
rna polymerase binding (20 genes)
proline-rich region binding (20 genes)
chromo shadow domain binding (5 genes)
fructose-6-phosphate binding (5 genes)
delta-catenin binding (9 genes)
sumo-specific endopeptidase activity (5 genes)
large ribosomal subunit rrna binding (7 genes)
small ribosomal subunit rrna binding (11 genes)
dna polymerase binding (18 genes)
phosphatidylinositol-4-phosphate binding (34 genes)
n-acylphosphatidylethanolamine-specific phospholipase d activity (6 genes)
phosphatidic acid binding (19 genes)
thyroid hormone binding (7 genes)
very-low-density lipoprotein particle receptor binding (4 genes)
aromatase activity (35 genes)
nadph binding (15 genes)
nad+ binding (14 genes)
nadh binding (4 genes)
co-smad binding (11 genes)
i-smad binding (11 genes)
r-smad binding (25 genes)
repressing transcription factor binding (40 genes)
oligosaccharide binding (13 genes)
death domain binding (5 genes)
k63-linked polyubiquitin modification-dependent protein binding (22 genes)
uridylyltransferase activity (5 genes)
lysine-acetylated histone binding (16 genes)
methylcytosine dioxygenase activity (4 genes)
proteasome binding (17 genes)
inositol 1,4,5 trisphosphate binding (16 genes)
bmp receptor binding (11 genes)
leucine binding (6 genes)
c2h2 zinc finger domain binding (14 genes)
dynein complex binding (24 genes)
growth factor receptor binding (4 genes)
primary mirna binding (9 genes)
pre-mirna binding (6 genes)
e-box binding (47 genes)
lipoteichoic acid binding (6 genes)
serine binding (4 genes)
eukaryotic initiation factor eif2 binding (5 genes)
connexin binding (7 genes)
ankyrin repeat binding (6 genes)
histone demethylase activity (h3-k27 specific) (5 genes)
linoleic acid epoxygenase activity (4 genes)
lipopeptide binding (7 genes)
chemokine (c-c motif) ligand 5 binding (4 genes)
lipoprotein particle binding (6 genes)
hmg box domain binding (25 genes)
14-3-3 protein binding (28 genes)
arp2/3 complex binding (11 genes)
fad binding (35 genes)
wd40-repeat domain binding (5 genes)
modified amino acid binding (7 genes)
naadp-sensitive calcium-release channel activity (5 genes)
protein phosphatase activator activity (8 genes)
phosphatidylinositol-3,5-bisphosphate binding (25 genes)
voltage-gated sodium channel activity involved in cardiac muscle cell action potential (5 genes)
protein binding involved in heterotypic cell-cell adhesion (6 genes)
caspase binding (7 genes)
vitamin transmembrane transporter activity (4 genes)
phosphatidylcholine-translocating atpase activity (4 genes)
receptor-receptor interaction (5 genes)
ceramide binding (5 genes)
l27 domain binding (4 genes)
ubiquitin-protein transferase activator activity (8 genes)
hedgehog family protein binding (4 genes)
neuroligin family protein binding (5 genes)
scaffold protein binding (76 genes)
cysteine-type endopeptidase activity involved in apoptotic process (10 genes)
pre-mrna intronic binding (9 genes)
mitochondrial ribosome binding (5 genes)
cysteine-type endopeptidase activity involved in apoptotic signaling pathway (6 genes)
cysteine-type endopeptidase activity involved in execution phase of apoptosis (4 genes)
7sk snrna binding (7 genes)
carbohydrate derivative binding (7 genes)
mdm2/mdm4 family protein binding (10 genes)
cyclin-dependent protein kinase activity (4 genes)
cullin family protein binding (18 genes)
amylin receptor activity (4 genes)
stat family protein binding (9 genes)
disordered domain specific binding (34 genes)
g-rich strand telomeric dna binding (11 genes)
nucleotide phosphatase activity, acting on free nucleotides (4 genes)
cell-cell adhesion mediator activity (20 genes)
collagen binding involved in cell-matrix adhesion (5 genes)
cadherin binding involved in cell-cell adhesion (5 genes)
molecular function regulator (4 genes)
bmp receptor activity (7 genes)
structural constituent of postsynaptic actin cytoskeleton (4 genes)
potassium channel activator activity (5 genes)
neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration (4 genes)
ubiquitinyl hydrolase activity (8 genes)
estrogen 16-alpha-hydroxylase activity (10 genes)
3-oxo-arachidoyl-coa synthase activity (7 genes)
3-oxo-cerotoyl-coa synthase activity (7 genes)
3-oxo-lignoceronyl-coa synthase activity (7 genes)
3-hydroxy-arachidoyl-coa dehydratase activity (4 genes)
3-hydroxy-behenoyl-coa dehydratase activity (4 genes)
3-hydroxy-lignoceroyl-coa dehydratase activity (4 genes)
decanoate--coa ligase activity (8 genes)
myristoyl-coa hydrolase activity (10 genes)
intermembrane lipid transfer activity (5 genes)
ubiquitin-dependent protein binding (7 genes)
cardiolipin binding (9 genes)
phosphatidylinositol phosphate binding (10 genes)
voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization (8 genes)
ceramide 1-phosphate binding (6 genes)
ceramide 1-phosphate transporter activity (6 genes)
phosphatidylinositol bisphosphate binding (5 genes)
cuprous ion binding (6 genes)
s-adenosyl-l-methionine binding (8 genes)
bat3 complex binding (4 genes)
transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential (33 genes)
voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential (4 genes)
retromer complex binding (5 genes)
polysome binding (6 genes)
ganglioside gt1b binding (4 genes)
phosphatidic acid transporter activity (6 genes)
histone methyltransferase binding (8 genes)
steroid hormone binding (6 genes)
n6-methyladenosine-containing rna binding (7 genes)
keratin filament binding (6 genes)
lys48-specific deubiquitinase activity (14 genes)
ubiquitin-specific protease binding (15 genes)
protein antigen binding (11 genes)
extracellular matrix protein binding (6 genes)
u1 snrnp binding (8 genes)
transferrin receptor binding (8 genes)
3' overhang single-stranded dna endodeoxyribonuclease activity (4 genes)
mrna cds binding (5 genes)
arrestin family protein binding (9 genes)
protein tyrosine kinase binding (23 genes)
sequence-specific mrna binding (12 genes)
sequence-specific double-stranded dna binding (33 genes)
promoter-specific chromatin binding (52 genes)
ubiquitin ligase inhibitor activity (6 genes)